For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKLSTATRRLLIGRPFRSDRLSHTLLPKRIALPVFASDALSSVAYAPEEVFLMLSVAGVSAYALTPWIGL AVAAVMLVVVASYRQNVHAYPSGGGDYEVVTTNLGETAGLVVASALMVDYVLTVAVSTASAMANIGSAVP IVAEHKVFFCVAAILLVMALNLRGVRESGVAFAIPTYAFIIGVLTMIGWGLFRIFVLGNPLRAESAGFEM HAQHGQVVGFALAFLVARSFSSGCAALTGVEAISNGVPAFQKPKSRNAATTLLMLGGIAVTLLMGIIVLA EKIGVKLVEDPATQLSGTPPGYYQKTLVTQLAQAVFGGFHIGFLLIATVTALILVLAANTAFNGFPVLGS ILAQHSYLPRQLHTRGDRLAFSNGILFLSGAALLAIIAFRGEVTALIQLYIVGVFISFTLSQIGMVRHWT RLLRTETDPAARRKMMRSRAVNTVGLLSTGAVLLVVLITKFAAGAWIALFAMSLLFGIMKMINRHYNTVN RELAEQAADQDDVVLPSRNHAVVLVSKLHLPTLRALAYARATRPDVLEAVTVSVDDAETRDLVHKWEEAD ISVPLKVIASPYREITRPVLEYVKRAREESPRTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFMPGV MVTSVPWQLSSSERRKTLEPHMAPGDARRGIFD*
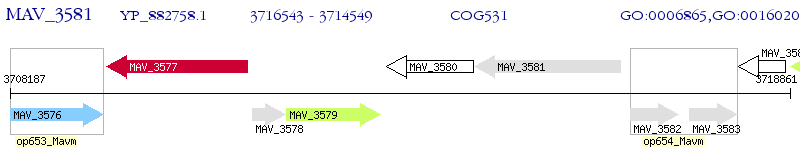
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3581 | - | - | 100% (664) | amino acid permease |
| M. avium 104 | MAV_1318 | - | 2e-98 | 33.70% (644) | OB-fold nucleic acid binding domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2709c | - | 0.0 | 82.08% (664) | hypothetical protein Mb2709c |
| M. gilvum PYR-GCK | Mflv_3926 | - | 0.0 | 77.41% (664) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv2690c | - | 0.0 | 82.04% (657) | hypothetical protein Rv2690c |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 9e-05 | 21.67% (240) | putative L-asparagine permease |
| M. abscessus ATCC 19977 | MAB_2993c | - | 0.0 | 79.12% (661) | hypothetical protein MAB_2993c |
| M. marinum M | MMAR_2024 | - | 0.0 | 83.61% (665) | integral membrane alanine, valine and leucine rich protein |
| M. smegmatis MC2 155 | MSMEG_2772 | - | 0.0 | 78.92% (664) | amino acid permease |
| M. thermoresistible (build 8) | TH_2211 | - | 0.0 | 74.44% (665) | PROBABLE CONSERVED INTEGRAL MEMBRANE ALANINE AND VALINE AND |
| M. ulcerans Agy99 | MUL_3328 | - | 0.0 | 83.46% (665) | integral membrane alanine, valine and leucine rich protein |
| M. vanbaalenii PYR-1 | Mvan_2474 | - | 0.0 | 77.11% (664) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2709c|M.bovis_AF2122/97 -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
Rv2690c|M.tuberculosis_H37Rv -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
MMAR_2024|M.marinum_M -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
MUL_3328|M.ulcerans_Agy99 -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
MAV_3581|M.avium_104 -------MSKLSTATRRLLIGRPFRSDRLSHTLLPKRIALPVFASDALSS
MAB_2993c|M.abscessus_ATCC_199 MLAFVSTLSKLSTAARRLVIGRPFRSDSLGHTLLPKRIALPVFASDALSS
Mflv_3926|M.gilvum_PYR-GCK -------MSKLSTAARRLVLGRPFRSDKLSHTLLPKRIALPVFASDALSS
Mvan_2474|M.vanbaalenii_PYR-1 -------MSKLSTVARRLVLGRPFRSDRLSHTLLPKRIALPVFASDALSS
MSMEG_2772|M.smegmatis_MC2_155 -------MSKLSTAARRLVIGRPFRSDKLSHTLLPKRIALPVFASDALSS
TH_2211|M.thermoresistible__bu -------VSKVSTAARRLLLGRPFRSDRLSHTLLPKRIALPVFASDALSS
MLBr_01305|M.leprae_Br4923 ----------MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGI
::* :. : : .* : :.
Mb2709c|M.bovis_AF2122/97 IAYAPEEIFLVLSVAGLAAYSMAPLIGLAVAAVLLVVVSSYRQNVHAYPS
Rv2690c|M.tuberculosis_H37Rv IAYAPEEIFLVLSVAGLAAYSMAPLIGLAVAAVLLVVVSSYRQNVHAYPS
MMAR_2024|M.marinum_M VAYAPEEIFLMLSVAGLAAYSMAPWIALAVAAVLLVVVSSYRQNVHAYPS
MUL_3328|M.ulcerans_Agy99 VAYAPEEIFLMLSVAGLAAYSMAPWIALAVAAVLLVVVSSYRQNVHAYPS
MAV_3581|M.avium_104 VAYAPEEVFLMLSVAGVSAYALTPWIGLAVAAVMLVVVASYRQNVHAYPS
MAB_2993c|M.abscessus_ATCC_199 VAYAPEEIFLVLSVAGMSAYAMTPWIGLAVAAVMMVVVASYRQNVHAYPS
Mflv_3926|M.gilvum_PYR-GCK TAYAPEEIFLVLSVAGLASYSMAPWIGLAVAAVMLIVIASYRQNVHAYPS
Mvan_2474|M.vanbaalenii_PYR-1 VAYAPEEIFLVLSVAGLAAYSMAPWIGLAVAGVMIIVIASYRQNVHAYPS
MSMEG_2772|M.smegmatis_MC2_155 VAYAPEEIFLVLSVAGLSAYTMTPWIGLAVAAVMLIVIASYRQNVHAYPS
TH_2211|M.thermoresistible__bu VAYAPEEIFVVLSVAGVAAYAMTPWIGLAIATVMALVIASYRHTVQAYPS
MLBr_01305|M.leprae_Br4923 GGAIGTGLFLGAGGRLAKAGPGLFLVYAVCGVFVFLILRALGELVLHRPS
. :*: . : . : . . .: ::: : . * **
Mb2709c|M.bovis_AF2122/97 GGGDYEVVTTNLGATGGLVVASALMVDYVLTVAVSISSAASNIGSVSPFV
Rv2690c|M.tuberculosis_H37Rv GGGDYEVVTTNLGATGGLVVASALMVDYVLTVAVSISSAASNIGSVSPFV
MMAR_2024|M.marinum_M GGGDYEVVSTNLGPTAGLVVASALMVDYVLTVAVSIASAMSNIGSAIPFV
MUL_3328|M.ulcerans_Agy99 GGGDYEVVSTNLGPTAGLVVASALMVDYVLTVAVSIASAMSNIGSAIPFV
MAV_3581|M.avium_104 GGGDYEVVTTNLGETAGLVVASALMVDYVLTVAVSTASAMANIGSAVPIV
MAB_2993c|M.abscessus_ATCC_199 GGGDYEVVTTNLGPTAGLTVGSALLVDYVLTVAVSMSSAMSNIGSAIPLV
Mflv_3926|M.gilvum_PYR-GCK GGGDYEIATTNLGPTAGLTVASALLVDYVLTVAVSMSSAMANIGSAVPFI
Mvan_2474|M.vanbaalenii_PYR-1 GGGDYEVVTTNLGPTAGLTVASALLIDYVLTVAVSMSSAMTNIGSAVPFI
MSMEG_2772|M.smegmatis_MC2_155 GGGDYEVVTTNLGPTAGLTVASALLVDYVLTVAVSMSSAMSNIGSAIPFV
TH_2211|M.thermoresistible__bu GGGDYEVVSTNLGRTAGLTVAAALMVDYVLTVAVSISAAMSNIGSVIGFI
MLBr_01305|M.leprae_Br4923 SGSFVSYAREFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFT
.*. . . :* ... .*. ::*:.:*. *. :: : : :
Mb2709c|M.bovis_AF2122/97 YEHKVLFAVGAIVLIMAMNLRGVRESGLAFAIPTYAFIAGIGTMLVWGLF
Rv2690c|M.tuberculosis_H37Rv YEHKVLFAVGAIVLIMAMNLRGVRESGLAFAIPTYAFIAGIGTMLVWGLF
MMAR_2024|M.marinum_M YDHKVFFAVAAILLVTALNLRGVRESGLAFAIPTYAFIIGISAMLGWGLF
MUL_3328|M.ulcerans_Agy99 YDHKVFFAVAAILLVTALNLRGVRESGLAFAIPTYAFIIGISAMLGWGLF
MAV_3581|M.avium_104 AEHKVFFCVAAILLVMALNLRGVRESGVAFAIPTYAFIIGVLTMIGWGLF
MAB_2993c|M.abscessus_ATCC_199 AQHKVTFAVAAIVILMSMNLRGVRESGAAFAIPTYAFMIGMYLMLGWGLF
Mflv_3926|M.gilvum_PYR-GCK NQHKVWFAVAAILVLAALNLRGIRESGTAFAIPTYAFIVGMSVMLIWGFV
Mvan_2474|M.vanbaalenii_PYR-1 NDHKVWFAVIAILVLASLNLRGIRESGTAFAIPTYAFMIGMYAMLAWGFL
MSMEG_2772|M.smegmatis_MC2_155 GHHKVLFAVAAILLLAAMNLRGIRESGVAFAIPTYLFMAGMFIMLGWGLF
TH_2211|M.thermoresistible__bu GEHKVWFAVAAILLLAAMNLRGVRESGTAFAVPTYVFVFAMVVMLGWGLI
MLBr_01305|M.leprae_Br4923 ALPQWTLALLALAVVLVMNLISVEWFG---ELEFWAALIKVCALMAFLVV
: :.: *: :: :** .:. * : : : : :: : ..
Mb2709c|M.bovis_AF2122/97 RIFVLG--NPVRAESAAFEMHAEHGQIVGFALVFLVARSFSSGCAALTGV
Rv2690c|M.tuberculosis_H37Rv RIFVLG--NPVRAESAAFEMHAEHGQIVGFALVFLVARSFSSGCAALTGV
MMAR_2024|M.marinum_M RIYVLG--NQVRAESAGFVMHAEHGKVVGIALVFLVARSFSSGCAALTGV
MUL_3328|M.ulcerans_Agy99 RIYVLG--NQVRAESAGFVMHAEHGKVVGIALVFLVARSFSSGCAALTGV
MAV_3581|M.avium_104 RIFVLG--NPLRAESAGFEMHAQHGQVVGFALAFLVARSFSSGCAALTGV
MAB_2993c|M.abscessus_ATCC_199 QIYVLG--TPLRAESASFEMHGEGNGILGFALVFLVARAFSSGCAALTGV
Mflv_3926|M.gilvum_PYR-GCK QIYVLG--TPLRAESADFEMHSEHGDVLGFALIFLVARAFSSGSAALTGV
Mvan_2474|M.vanbaalenii_PYR-1 QILVLG--RDLRAESAGFEMHSEHGEVLGFALVFLVARAFSSGSAALTGV
MSMEG_2772|M.smegmatis_MC2_155 QIYVLD--HSLRAESADFQMHSEHGEVLGFALVFLVARAFSSGSAALTGV
TH_2211|M.thermoresistible__bu RIYLFD--TPLRAESAAFEMIPPDGEIVGFALVFLVARAFSSGSAALTGI
MLBr_01305|M.leprae_Br4923 GTIFLGGRYPVDGHNTGLSLWTSHGGLFPTGVAPLIVVSSG-VMFAYAAV
.:. : ...: : : . :. .: *:. : . * :.:
Mb2709c|M.bovis_AF2122/97 EAISNGVPAFQKPKSRNAATTLLMLGIIAVSMFMGMIVLAVETGVQVVDD
Rv2690c|M.tuberculosis_H37Rv EAISNGVPAFQKPKSRNAATTLLMLGIIAVSMFMGMIVLAVETGVQVVDD
MMAR_2024|M.marinum_M EAISNGVPAFQKPKSRNAATTLLMLGAIAVALLMGVVALAVQTGAQVVDD
MUL_3328|M.ulcerans_Agy99 EAISNGVPAFQKPKSRNAATTLLMLGAIAVALLMGVVALAVQTGAQVVDD
MAV_3581|M.avium_104 EAISNGVPAFQKPKSRNAATTLLMLGGIAVTLLMGIIVLAEKIGVKLVED
MAB_2993c|M.abscessus_ATCC_199 EAISNGVPAFRKPKSRNAATTLLLLGGIAITLFMGIILLAERTGAKIAED
Mflv_3926|M.gilvum_PYR-GCK EAISNGVPAFRKPKSRNAATTLLLLGLIATTLLMGIIMLARAIGVQIAER
Mvan_2474|M.vanbaalenii_PYR-1 EAISNGVPAFRKPKSRNAATTLLLLGVIAITLFMGMILLARATGVQVAER
MSMEG_2772|M.smegmatis_MC2_155 EAISNGVPAFRKPKSRNAATTLLLLGMIAVTLFMGIILLAKATGVQIAEH
TH_2211|M.thermoresistible__bu EAISNGVPAFRKPKARNAATTLLVLGLIAVPMFLGTVVLAERTGVQIAER
MLBr_01305|M.leprae_Br4923 ELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGSVILLALLLPYSAFKA
* :.... :** . : ::. ** . . : ** . .
Mb2709c|M.bovis_AF2122/97 PDTQLTGAPPGYQQKTLVAQLAQAVFGGFYLGFLLIAAVTALILVLAANT
Rv2690c|M.tuberculosis_H37Rv PDTQLTGAPPGYQQKTLVAQLAQAVFGGFYLGFLLIAAVTALILVLAANT
MMAR_2024|M.marinum_M PDTQLTGAPDGYQQKTLVAQLAQAVFGSFHLGFLMIAAVTALILVLAANT
MUL_3328|M.ulcerans_Agy99 PDTQLTGAPDGYQQKTLVAQLAQAVFGSFHLGFLMIAAVTALILVLAANT
MAV_3581|M.avium_104 PATQLSGTPPGYYQKTLVTQLAQAVFGGFHIGFLLIATVTALILVLAANT
MAB_2993c|M.abscessus_ATCC_199 PTRQLSGAPADYHQKTLVAQLAEAVFGDFRIGFLFITVVTALILVLAANT
Mflv_3926|M.gilvum_PYR-GCK PAEQLVGAPEDYHQKTLIAQLAETVFHGFPFGLFFIALVTALILVLAANT
Mvan_2474|M.vanbaalenii_PYR-1 PLEQLGGAPADYQQKTLIAQLADAVFHDFPLGLYLIAGVTALILVLAANT
MSMEG_2772|M.smegmatis_MC2_155 PHVQLIGAPTDYHQKTLIAQLADAVFHGFPLGLYLIATVTALILCLAANT
TH_2211|M.thermoresistible__bu PHAQFVGAPPDYQQKTLIAQLAGAVFDGWGPAVLATVIAAALILLLAANT
MLBr_01305|M.leprae_Br4923 S------------ESPFVTFFSKVGFYGAGDLMNIVVLTAALSSLNAGLY
. :..::: :: . * . . . .:** *.
Mb2709c|M.bovis_AF2122/97 AFNGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGILFLAAAAIGAVVAFR
Rv2690c|M.tuberculosis_H37Rv AFNGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGILFLAAAAIGAVVAFR
MMAR_2024|M.marinum_M AFNGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGIIFLSAAAVGAVVAFR
MUL_3328|M.ulcerans_Agy99 AFNGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGIIFLSAAAVGAVVAFR
MAV_3581|M.avium_104 AFNGFPVLGSILAQHSYLPRQLHTRGDRLAFSNGILFLSGAALLAIIAFR
MAB_2993c|M.abscessus_ATCC_199 AFNGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGIIFLSAVAILFVVAFK
Mflv_3926|M.gilvum_PYR-GCK AFNGFPVLGSILAQDRFLPRQLHTRGDRLAFSNGILFLAFGAVAFVVAFQ
Mvan_2474|M.vanbaalenii_PYR-1 AFNGFPVLGSILAQDRFLPRQLHTRGDRLAFSNGILFLAFGAIAFVVAFQ
MSMEG_2772|M.smegmatis_MC2_155 AFNGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGILILALAAIAFVVAFG
TH_2211|M.thermoresistible__bu AFNGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGILFLSLVASAFVVAFG
MLBr_01305|M.leprae_Br4923 ATG--RVMHSIAINGSGPKFTARMSKNGVPYG-GILLAAVICLCGVALNA
* . *: *: : : : :.:. **:: : . :
Mb2709c|M.bovis_AF2122/97 AELTALIQLYIVGVFISFTMSQVGMVRHWTRLLSAETDPRARRAMLRSRA
Rv2690c|M.tuberculosis_H37Rv AELTALIQLYIVGVFISFTMSQVGMVRHWTRLLSAETDPRARRAMLRSRA
MMAR_2024|M.marinum_M AELTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDVAARRKMVRSRV
MUL_3328|M.ulcerans_Agy99 AELTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDVAARRKMVRSRV
MAV_3581|M.avium_104 GEVTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDPAARRKMMRSRA
MAB_2993c|M.abscessus_ATCC_199 AEVTALIQLYIVGVFVSFTLSQIGMVRHWTRLLKTETDPAVRRKMMRSRV
Mflv_3926|M.gilvum_PYR-GCK AEVTKLIQLYIVGVFVSFTLSQIGMVRHWTRLLRTETDAAARGRMVRARV
Mvan_2474|M.vanbaalenii_PYR-1 AEVTALIQLYIVGVFVSFTFSQIGMVRHWTRLLRTETDPAARRRMMRARV
MSMEG_2772|M.smegmatis_MC2_155 AEVTALIHLYIVGVFVSFTFSQIGMVRHWTRLLRNETDPAIRRRMMRSRA
TH_2211|M.thermoresistible__bu AEVSALIQLYIVGVFVSFTFSQLGMVRHWTRSLRGETEPAVRRTMHRARA
MLBr_01305|M.leprae_Br4923 FNPGQAFEIVLSVAALGIIAGWGTIVLCQLRLHKMAKAGIMRRPRFRMPL
: :.: : . :.: . :* * . * *
Mb2709c|M.bovis_AF2122/97 VNTVGFVSTGTVLLIVLVTKFLAGAWIAIVAMGGFFMMMKLIHRHYDAVN
Rv2690c|M.tuberculosis_H37Rv VNTVGFVSTGTVLLIVLVTKFLAGAWIAIVAMGGFFMMMKLIHRHYDAVN
MMAR_2024|M.marinum_M VNSVGLVSTGAVLLIVLVTKFLAGAWIAIVAMGSVFILMKMIRRHYDTVN
MUL_3328|M.ulcerans_Agy99 VNSVGLVSTGAVLLIVLVTKFLAGAWIAIVAMGSVFILMKMIRRHYDTVN
MAV_3581|M.avium_104 VNTVGLLSTGAVLLVVLITKFAAGAWIALFAMSLLFGIMKMINRHYNTVN
MAB_2993c|M.abscessus_ATCC_199 INTIGFIMTGTVLLVVLVTKFLAGAWIAILAMGLLFVLMKMIHKHYNTVA
Mflv_3926|M.gilvum_PYR-GCK INTVGFLCTGTVLIIVVVTKFVVGAWIAILAMGALFGIMKLIHRHYASVS
Mvan_2474|M.vanbaalenii_PYR-1 INSVGLVCTGTVLVIVVVTKFLAGAWIAILAMSALFGLMKLIHKHYASVT
MSMEG_2772|M.smegmatis_MC2_155 INTVGLTATGAVLVVVVVTKFVAGAWIAILAMVALFVLMKLIHRHYDTVA
TH_2211|M.thermoresistible__bu VNAVGCVATGSVLLVVVVTKFAAGAWMAILTMAVLFMLMRLIHRHYATVA
MLBr_01305|M.leprae_Br4923 APYSGYLTLAFLFAVLVVMAFDK--PIGTWTVASLIVIVPALIAGWYSIR
* . :: :::: * :. :: .: :: : : ::
Mb2709c|M.bovis_AF2122/97 RELAEQAEEA--EITLPSRNHAVVLVSKLHLPTLRALTYARATRPDVLEA
Rv2690c|M.tuberculosis_H37Rv RELAEQAEEA--EITLPSRNHAVVLVSKLHLPTLRALTYARATRPDVLEA
MMAR_2024|M.marinum_M RELEEQAAAHDNEVVLPSRNHALVLVSKLHLPTLRALAYARATRPDVLEA
MUL_3328|M.ulcerans_Agy99 RELEEQAAAHGNEVVLPSRNHALVLVSKLHLPTLRALAYARATRPDVLEA
MAV_3581|M.avium_104 RELAEQAADQD-DVVLPSRNHAVVLVSKLHLPTLRALAYARATRPDVLEA
MAB_2993c|M.abscessus_ATCC_199 RELDN----HEGEVVLPSRTHAVVLVSKVHQPTLRALAYARATRPDVLEA
Mflv_3926|M.gilvum_PYR-GCK RELEARAADAE-DIVLPSRNHAVVLVSNLHMPTLRALAYARATRPDVLEA
Mvan_2474|M.vanbaalenii_PYR-1 RELEARAAETE-DIVLPSRNHAIVLVSNAHLPTLRALAYARATRPDVLEA
MSMEG_2772|M.smegmatis_MC2_155 RELEAQAEDED-DIVLPSRNHAVVLVSKLHLPTKRALAYARATRPDVLEA
TH_2211|M.thermoresistible__bu RELDERAAADDDGMVLPSRNHAIVLVSNLHLPTLRALAYARATRPDVLEA
MLBr_01305|M.leprae_Br4923 ---------------------------------KRVMTIARER-------
*.:: **
Mb2709c|M.bovis_AF2122/97 VTVNVDDAETRELVRQWQDSDVSVPLKVIASPYREITRPVLDYVKRVSKE
Rv2690c|M.tuberculosis_H37Rv VTVNVDDAETRELVRQWQDSDVSVPLKVIASPYREITRPVLDYVKRVSKE
MMAR_2024|M.marinum_M ITVSVDDVETRELVRQWEDSDVSVPLKVIASPYREITRPVLDYVKRVSKE
MUL_3328|M.ulcerans_Agy99 ITVSVDDVETRELVRQWEDSDVSVPLKVIASPYREITRPVLDYVKRVSKE
MAV_3581|M.avium_104 VTVSVDDAETRDLVHKWEEADISVPLKVIASPYREITRPVLEYVKRAREE
MAB_2993c|M.abscessus_ATCC_199 VTVSVDDRDTRELVREWETSDISTPLKVIASPYREITRPILDYVKRISKE
Mflv_3926|M.gilvum_PYR-GCK ITVSVDDAETRELVHKWENSDVSVPLKVIASPYREITRPVLDYVKRVTKD
Mvan_2474|M.vanbaalenii_PYR-1 ITVSVDDAETRQLVHKWEHSDISVPLKVVASPYREITRPVLDYVKRVGKD
MSMEG_2772|M.smegmatis_MC2_155 ITVSVDDAETRALVHEWEGSDVAVPLKVIASPYREITRPVLDYVKRITKE
TH_2211|M.thermoresistible__bu VTVSVDDAETRELVHKWEESDVTVPLKVIASPYREVTRPVLDYVKRVTKE
MLBr_01305|M.leprae_Br4923 --------------------------MGYTGPFPAIANPPVQPSERSHSQ
:.*: ::.* :: :* .:
Mb2709c|M.bovis_AF2122/97 SPRTVVTVFIPEYVVGRWWEQLLHNQSALRLKGRLLFMPGVMVTSVPWQL
Rv2690c|M.tuberculosis_H37Rv SPRTVVTVFIPEYVVGRWWEQLLHNQSALRLKGRLLFMPGVMVTSVPWQL
MMAR_2024|M.marinum_M SPRTVVTVFIPEYVVGRWWEQLLHNQSAFRLKTRLLFMPGVMVTSVPWQL
MUL_3328|M.ulcerans_Agy99 SPRTVVTVFIPEYVVGRWWEQLLHNQSAFRLKTRLLFMPGVMVTSVPWQL
MAV_3581|M.avium_104 SPRTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFMPGVMVTSVPWQL
MAB_2993c|M.abscessus_ATCC_199 APRTVVCIFIPEYVVGHWWEQVLHNQSALRLKGRLLFVPNVMVTSVPWQL
Mflv_3926|M.gilvum_PYR-GCK SPRTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFEPNVMVTSVPWQL
Mvan_2474|M.vanbaalenii_PYR-1 SPRTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFEPNVMVTSVPWQL
MSMEG_2772|M.smegmatis_MC2_155 SPRTVVTVFIPEYVVGHWWEQILHNQSALRLKGRLLFMPNVMVTSVPWQL
TH_2211|M.thermoresistible__bu SPRTVVTVFIPEYVVGHWWEQLLHNQSALRLKGRLLFMPNVMVTSVPWQL
MLBr_01305|M.leprae_Br4923 NP------------------------------------------------
*
Mb2709c|M.bovis_AF2122/97 TSSERIKTLQPHAAPGDTRRGIFD
Rv2690c|M.tuberculosis_H37Rv TSSERIKTLQPHAAPGDT------
MMAR_2024|M.marinum_M TSSERINTLEPHAAPGDMRRGIFD
MUL_3328|M.ulcerans_Agy99 TSSERINTLEPHAAPGDTRRGIFD
MAV_3581|M.avium_104 SSSERRKTLEPHMAPGDARRGIFD
MAB_2993c|M.abscessus_ATCC_199 DSSERRRELDRQSAPGDSRRGFDS
Mflv_3926|M.gilvum_PYR-GCK TSSERLKKMAPQSAPGDARRGFLE
Mvan_2474|M.vanbaalenii_PYR-1 SSSERLKKLAPQSAPGDARRGFLD
MSMEG_2772|M.smegmatis_MC2_155 TSSERLKAIQRQSAPGDARRGFLE
TH_2211|M.thermoresistible__bu SSSERRRKLQPQWAPGDARRGFLE
MLBr_01305|M.leprae_Br4923 ------------------------