For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSKLSTAARRLVIGRPFRSDKLSHTLLPKRIALPVFASDALSSVAYAPEEIFLVLSVAGLSAYTMTPWIG LAVAAVMLIVIASYRQNVHAYPSGGGDYEVVTTNLGPTAGLTVASALLVDYVLTVAVSMSSAMSNIGSAI PFVGHHKVLFAVAAILLLAAMNLRGIRESGVAFAIPTYLFMAGMFIMLGWGLFQIYVLDHSLRAESADFQ MHSEHGEVLGFALVFLVARAFSSGSAALTGVEAISNGVPAFRKPKSRNAATTLLLLGMIAVTLFMGIILL AKATGVQIAEHPHVQLIGAPTDYHQKTLIAQLADAVFHGFPLGLYLIATVTALILCLAANTAFNGFPVLG SILAQDRYLPRQLHTRGDRLAFSNGILILALAAIAFVVAFGAEVTALIHLYIVGVFVSFTFSQIGMVRHW TRLLRNETDPAIRRRMMRSRAINTVGLTATGAVLVVVVVTKFVAGAWIAILAMVALFVLMKLIHRHYDTV ARELEAQAEDEDDIVLPSRNHAVVLVSKLHLPTKRALAYARATRPDVLEAITVSVDDAETRALVHEWEGS DVAVPLKVIASPYREITRPVLDYVKRITKESPRTVVTVFIPEYVVGHWWEQILHNQSALRLKGRLLFMPN VMVTSVPWQLTSSERLKAIQRQSAPGDARRGFLE
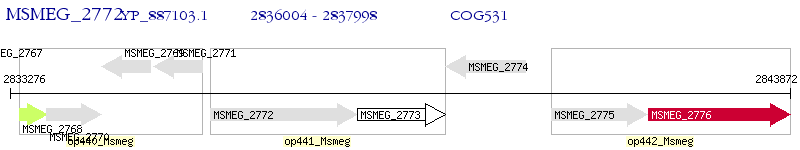
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2772 | - | - | 100% (664) | amino acid permease |
| M. smegmatis MC2 155 | MSMEG_1552 | eat | 3e-09 | 20.79% (457) | ethanolamine permease |
| M. smegmatis MC2 155 | MSMEG_6727 | - | 9e-07 | 20.05% (429) | amino acid permease-associated region |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2709c | - | 0.0 | 78.61% (664) | hypothetical protein Mb2709c |
| M. gilvum PYR-GCK | Mflv_3926 | - | 0.0 | 85.39% (664) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv2690c | - | 0.0 | 79.00% (657) | hypothetical protein Rv2690c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2993c | - | 0.0 | 81.87% (662) | hypothetical protein MAB_2993c |
| M. marinum M | MMAR_2024 | - | 0.0 | 78.05% (665) | integral membrane alanine, valine and leucine rich protein |
| M. avium 104 | MAV_3581 | - | 0.0 | 78.92% (664) | amino acid permease |
| M. thermoresistible (build 8) | TH_2211 | - | 0.0 | 79.70% (665) | PROBABLE CONSERVED INTEGRAL MEMBRANE ALANINE AND VALINE AND |
| M. ulcerans Agy99 | MUL_3328 | - | 0.0 | 77.89% (665) | integral membrane alanine, valine and leucine rich protein |
| M. vanbaalenii PYR-1 | Mvan_2474 | - | 0.0 | 85.69% (664) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3926|M.gilvum_PYR-GCK -------MSKLSTAARRLVLGRPFRSDKLSHTLLPKRIALPVFASDALSS
Mvan_2474|M.vanbaalenii_PYR-1 -------MSKLSTVARRLVLGRPFRSDRLSHTLLPKRIALPVFASDALSS
MSMEG_2772|M.smegmatis_MC2_155 -------MSKLSTAARRLVIGRPFRSDKLSHTLLPKRIALPVFASDALSS
TH_2211|M.thermoresistible__bu -------VSKVSTAARRLLLGRPFRSDRLSHTLLPKRIALPVFASDALSS
MAB_2993c|M.abscessus_ATCC_199 MLAFVSTLSKLSTAARRLVIGRPFRSDSLGHTLLPKRIALPVFASDALSS
Mb2709c|M.bovis_AF2122/97 -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
Rv2690c|M.tuberculosis_H37Rv -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
MMAR_2024|M.marinum_M -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
MUL_3328|M.ulcerans_Agy99 -------MSKLSTAARRLLIGRPFRSDRLSHTLLPKRIALPVFASDAMSS
MAV_3581|M.avium_104 -------MSKLSTATRRLLIGRPFRSDRLSHTLLPKRIALPVFASDALSS
:**:**.:***::******* *.*****************:**
Mflv_3926|M.gilvum_PYR-GCK TAYAPEEIFLVLSVAGLASYSMAPWIGLAVAAVMLIVIASYRQNVHAYPS
Mvan_2474|M.vanbaalenii_PYR-1 VAYAPEEIFLVLSVAGLAAYSMAPWIGLAVAGVMIIVIASYRQNVHAYPS
MSMEG_2772|M.smegmatis_MC2_155 VAYAPEEIFLVLSVAGLSAYTMTPWIGLAVAAVMLIVIASYRQNVHAYPS
TH_2211|M.thermoresistible__bu VAYAPEEIFVVLSVAGVAAYAMTPWIGLAIATVMALVIASYRHTVQAYPS
MAB_2993c|M.abscessus_ATCC_199 VAYAPEEIFLVLSVAGMSAYAMTPWIGLAVAAVMMVVVASYRQNVHAYPS
Mb2709c|M.bovis_AF2122/97 IAYAPEEIFLVLSVAGLAAYSMAPLIGLAVAAVLLVVVSSYRQNVHAYPS
Rv2690c|M.tuberculosis_H37Rv IAYAPEEIFLVLSVAGLAAYSMAPLIGLAVAAVLLVVVSSYRQNVHAYPS
MMAR_2024|M.marinum_M VAYAPEEIFLMLSVAGLAAYSMAPWIALAVAAVLLVVVSSYRQNVHAYPS
MUL_3328|M.ulcerans_Agy99 VAYAPEEIFLMLSVAGLAAYSMAPWIALAVAAVLLVVVSSYRQNVHAYPS
MAV_3581|M.avium_104 VAYAPEEVFLMLSVAGVSAYALTPWIGLAVAAVMLVVVASYRQNVHAYPS
******:*::*****:::*:::* *.**:* *: :*::***:.*:****
Mflv_3926|M.gilvum_PYR-GCK GGGDYEIATTNLGPTAGLTVASALLVDYVLTVAVSMSSAMANIGSAVPFI
Mvan_2474|M.vanbaalenii_PYR-1 GGGDYEVVTTNLGPTAGLTVASALLIDYVLTVAVSMSSAMTNIGSAVPFI
MSMEG_2772|M.smegmatis_MC2_155 GGGDYEVVTTNLGPTAGLTVASALLVDYVLTVAVSMSSAMSNIGSAIPFV
TH_2211|M.thermoresistible__bu GGGDYEVVSTNLGRTAGLTVAAALMVDYVLTVAVSISAAMSNIGSVIGFI
MAB_2993c|M.abscessus_ATCC_199 GGGDYEVVTTNLGPTAGLTVGSALLVDYVLTVAVSMSSAMSNIGSAIPLV
Mb2709c|M.bovis_AF2122/97 GGGDYEVVTTNLGATGGLVVASALMVDYVLTVAVSISSAASNIGSVSPFV
Rv2690c|M.tuberculosis_H37Rv GGGDYEVVTTNLGATGGLVVASALMVDYVLTVAVSISSAASNIGSVSPFV
MMAR_2024|M.marinum_M GGGDYEVVSTNLGPTAGLVVASALMVDYVLTVAVSIASAMSNIGSAIPFV
MUL_3328|M.ulcerans_Agy99 GGGDYEVVSTNLGPTAGLVVASALMVDYVLTVAVSIASAMSNIGSAIPFV
MAV_3581|M.avium_104 GGGDYEVVTTNLGETAGLVVASALMVDYVLTVAVSTASAMANIGSAVPIV
******:.:**** *.**.*.:**::********* ::* :****. ::
Mflv_3926|M.gilvum_PYR-GCK NQHKVWFAVAAILVLAALNLRGIRESGTAFAIPTYAFIVGMSVMLIWGFV
Mvan_2474|M.vanbaalenii_PYR-1 NDHKVWFAVIAILVLASLNLRGIRESGTAFAIPTYAFMIGMYAMLAWGFL
MSMEG_2772|M.smegmatis_MC2_155 GHHKVLFAVAAILLLAAMNLRGIRESGVAFAIPTYLFMAGMFIMLGWGLF
TH_2211|M.thermoresistible__bu GEHKVWFAVAAILLLAAMNLRGVRESGTAFAVPTYVFVFAMVVMLGWGLI
MAB_2993c|M.abscessus_ATCC_199 AQHKVTFAVAAIVILMSMNLRGVRESGAAFAIPTYAFMIGMYLMLGWGLF
Mb2709c|M.bovis_AF2122/97 YEHKVLFAVGAIVLIMAMNLRGVRESGLAFAIPTYAFIAGIGTMLVWGLF
Rv2690c|M.tuberculosis_H37Rv YEHKVLFAVGAIVLIMAMNLRGVRESGLAFAIPTYAFIAGIGTMLVWGLF
MMAR_2024|M.marinum_M YDHKVFFAVAAILLVTALNLRGVRESGLAFAIPTYAFIIGISAMLGWGLF
MUL_3328|M.ulcerans_Agy99 YDHKVFFAVAAILLVTALNLRGVRESGLAFAIPTYAFIIGISAMLGWGLF
MAV_3581|M.avium_104 AEHKVFFCVAAILLVMALNLRGVRESGVAFAIPTYAFIIGVLTMIGWGLF
.*** *.* **::: ::****:**** ***:*** *: .: *: **:.
Mflv_3926|M.gilvum_PYR-GCK QIYVLGTPLRAESADFEMHSEHGDVLGFALIFLVARAFSSGSAALTGVEA
Mvan_2474|M.vanbaalenii_PYR-1 QILVLGRDLRAESAGFEMHSEHGEVLGFALVFLVARAFSSGSAALTGVEA
MSMEG_2772|M.smegmatis_MC2_155 QIYVLDHSLRAESADFQMHSEHGEVLGFALVFLVARAFSSGSAALTGVEA
TH_2211|M.thermoresistible__bu RIYLFDTPLRAESAAFEMIPPDGEIVGFALVFLVARAFSSGSAALTGIEA
MAB_2993c|M.abscessus_ATCC_199 QIYVLGTPLRAESASFEMHGEGNGILGFALVFLVARAFSSGCAALTGVEA
Mb2709c|M.bovis_AF2122/97 RIFVLGNPVRAESAAFEMHAEHGQIVGFALVFLVARSFSSGCAALTGVEA
Rv2690c|M.tuberculosis_H37Rv RIFVLGNPVRAESAAFEMHAEHGQIVGFALVFLVARSFSSGCAALTGVEA
MMAR_2024|M.marinum_M RIYVLGNQVRAESAGFVMHAEHGKVVGIALVFLVARSFSSGCAALTGVEA
MUL_3328|M.ulcerans_Agy99 RIYVLGNQVRAESAGFVMHAEHGKVVGIALVFLVARSFSSGCAALTGVEA
MAV_3581|M.avium_104 RIFVLGNPLRAESAGFEMHAQHGQVVGFALAFLVARSFSSGCAALTGVEA
:* ::. :***** * * . ::*:** *****:****.*****:**
Mflv_3926|M.gilvum_PYR-GCK ISNGVPAFRKPKSRNAATTLLLLGLIATTLLMGIIMLARAIGVQIAERPA
Mvan_2474|M.vanbaalenii_PYR-1 ISNGVPAFRKPKSRNAATTLLLLGVIAITLFMGMILLARATGVQVAERPL
MSMEG_2772|M.smegmatis_MC2_155 ISNGVPAFRKPKSRNAATTLLLLGMIAVTLFMGIILLAKATGVQIAEHPH
TH_2211|M.thermoresistible__bu ISNGVPAFRKPKARNAATTLLVLGLIAVPMFLGTVVLAERTGVQIAERPH
MAB_2993c|M.abscessus_ATCC_199 ISNGVPAFRKPKSRNAATTLLLLGGIAITLFMGIILLAERTGAKIAEDPT
Mb2709c|M.bovis_AF2122/97 ISNGVPAFQKPKSRNAATTLLMLGIIAVSMFMGMIVLAVETGVQVVDDPD
Rv2690c|M.tuberculosis_H37Rv ISNGVPAFQKPKSRNAATTLLMLGIIAVSMFMGMIVLAVETGVQVVDDPD
MMAR_2024|M.marinum_M ISNGVPAFQKPKSRNAATTLLMLGAIAVALLMGVVALAVQTGAQVVDDPD
MUL_3328|M.ulcerans_Agy99 ISNGVPAFQKPKSRNAATTLLMLGAIAVALLMGVVALAVQTGAQVVDDPD
MAV_3581|M.avium_104 ISNGVPAFQKPKSRNAATTLLMLGGIAVTLLMGIIVLAEKIGVKLVEDPA
********:***:********:** ** .:::* : ** *.::.: *
Mflv_3926|M.gilvum_PYR-GCK EQLVGAPEDYHQKTLIAQLAETVFHGFPFGLFFIALVTALILVLAANTAF
Mvan_2474|M.vanbaalenii_PYR-1 EQLGGAPADYQQKTLIAQLADAVFHDFPLGLYLIAGVTALILVLAANTAF
MSMEG_2772|M.smegmatis_MC2_155 VQLIGAPTDYHQKTLIAQLADAVFHGFPLGLYLIATVTALILCLAANTAF
TH_2211|M.thermoresistible__bu AQFVGAPPDYQQKTLIAQLAGAVFDGWGPAVLATVIAAALILLLAANTAF
MAB_2993c|M.abscessus_ATCC_199 RQLSGAPADYHQKTLVAQLAEAVFGDFRIGFLFITVVTALILVLAANTAF
Mb2709c|M.bovis_AF2122/97 TQLTGAPPGYQQKTLVAQLAQAVFGGFYLGFLLIAAVTALILVLAANTAF
Rv2690c|M.tuberculosis_H37Rv TQLTGAPPGYQQKTLVAQLAQAVFGGFYLGFLLIAAVTALILVLAANTAF
MMAR_2024|M.marinum_M TQLTGAPDGYQQKTLVAQLAQAVFGSFHLGFLMIAAVTALILVLAANTAF
MUL_3328|M.ulcerans_Agy99 TQLTGAPDGYQQKTLVAQLAQAVFGSFHLGFLMIAAVTALILVLAANTAF
MAV_3581|M.avium_104 TQLSGTPPGYYQKTLVTQLAQAVFGGFHIGFLLIATVTALILVLAANTAF
*: *:* .* ****::*** :** .: .. . .:**** *******
Mflv_3926|M.gilvum_PYR-GCK NGFPVLGSILAQDRFLPRQLHTRGDRLAFSNGILFLAFGAVAFVVAFQAE
Mvan_2474|M.vanbaalenii_PYR-1 NGFPVLGSILAQDRFLPRQLHTRGDRLAFSNGILFLAFGAIAFVVAFQAE
MSMEG_2772|M.smegmatis_MC2_155 NGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGILILALAAIAFVVAFGAE
TH_2211|M.thermoresistible__bu NGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGILFLSLVASAFVVAFGAE
MAB_2993c|M.abscessus_ATCC_199 NGFPVLGSILAQDRYLPRQLHTRGDRLAFSNGIIFLSAVAILFVVAFKAE
Mb2709c|M.bovis_AF2122/97 NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGILFLAAAAIGAVVAFRAE
Rv2690c|M.tuberculosis_H37Rv NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGILFLAAAAIGAVVAFRAE
MMAR_2024|M.marinum_M NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGIIFLSAAAVGAVVAFRAE
MUL_3328|M.ulcerans_Agy99 NGFPVLGSVLAQHSYLPRQLHTRGDRLAFSNGIIFLSAAAVGAVVAFRAE
MAV_3581|M.avium_104 NGFPVLGSILAQHSYLPRQLHTRGDRLAFSNGILFLSGAALLAIIAFRGE
********:***. :******************::*: * ::** .*
Mflv_3926|M.gilvum_PYR-GCK VTKLIQLYIVGVFVSFTLSQIGMVRHWTRLLRTETDAAARGRMVRARVIN
Mvan_2474|M.vanbaalenii_PYR-1 VTALIQLYIVGVFVSFTFSQIGMVRHWTRLLRTETDPAARRRMMRARVIN
MSMEG_2772|M.smegmatis_MC2_155 VTALIHLYIVGVFVSFTFSQIGMVRHWTRLLRNETDPAIRRRMMRSRAIN
TH_2211|M.thermoresistible__bu VSALIQLYIVGVFVSFTFSQLGMVRHWTRSLRGETEPAVRRTMHRARAVN
MAB_2993c|M.abscessus_ATCC_199 VTALIQLYIVGVFVSFTLSQIGMVRHWTRLLKTETDPAVRRKMMRSRVIN
Mb2709c|M.bovis_AF2122/97 LTALIQLYIVGVFISFTMSQVGMVRHWTRLLSAETDPRARRAMLRSRAVN
Rv2690c|M.tuberculosis_H37Rv LTALIQLYIVGVFISFTMSQVGMVRHWTRLLSAETDPRARRAMLRSRAVN
MMAR_2024|M.marinum_M LTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDVAARRKMVRSRVVN
MUL_3328|M.ulcerans_Agy99 LTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDVAARRKMVRSRVVN
MAV_3581|M.avium_104 VTALIQLYIVGVFISFTLSQIGMVRHWTRLLRTETDPAARRKMMRSRAVN
:: **:*******:***:**:******** * **: * * *:*.:*
Mflv_3926|M.gilvum_PYR-GCK TVGFLCTGTVLIIVVVTKFVVGAWIAILAMGALFGIMKLIHRHYASVSRE
Mvan_2474|M.vanbaalenii_PYR-1 SVGLVCTGTVLVIVVVTKFLAGAWIAILAMSALFGLMKLIHKHYASVTRE
MSMEG_2772|M.smegmatis_MC2_155 TVGLTATGAVLVVVVVTKFVAGAWIAILAMVALFVLMKLIHRHYDTVARE
TH_2211|M.thermoresistible__bu AVGCVATGSVLLVVVVTKFAAGAWMAILTMAVLFMLMRLIHRHYATVARE
MAB_2993c|M.abscessus_ATCC_199 TIGFIMTGTVLLVVLVTKFLAGAWIAILAMGLLFVLMKMIHKHYNTVARE
Mb2709c|M.bovis_AF2122/97 TVGFVSTGTVLLIVLVTKFLAGAWIAIVAMGGFFMMMKLIHRHYDAVNRE
Rv2690c|M.tuberculosis_H37Rv TVGFVSTGTVLLIVLVTKFLAGAWIAIVAMGGFFMMMKLIHRHYDAVNRE
MMAR_2024|M.marinum_M SVGLVSTGAVLLIVLVTKFLAGAWIAIVAMGSVFILMKMIRRHYDTVNRE
MUL_3328|M.ulcerans_Agy99 SVGLVSTGAVLLIVLVTKFLAGAWIAIVAMGSVFILMKMIRRHYDTVNRE
MAV_3581|M.avium_104 TVGLLSTGAVLLVVLITKFAAGAWIALFAMSLLFGIMKMINRHYNTVNRE
::* **:**::*::*** .***:*:.:* .* :*::*.:** :* **
Mflv_3926|M.gilvum_PYR-GCK LEARAADAED-IVLPSRNHAVVLVSNLHMPTLRALAYARATRPDVLEAIT
Mvan_2474|M.vanbaalenii_PYR-1 LEARAAETED-IVLPSRNHAIVLVSNAHLPTLRALAYARATRPDVLEAIT
MSMEG_2772|M.smegmatis_MC2_155 LEAQAEDEDD-IVLPSRNHAVVLVSKLHLPTKRALAYARATRPDVLEAIT
TH_2211|M.thermoresistible__bu LDERAAADDDGMVLPSRNHAIVLVSNLHLPTLRALAYARATRPDVLEAVT
MAB_2993c|M.abscessus_ATCC_199 LDN----HEGEVVLPSRTHAVVLVSKVHQPTLRALAYARATRPDVLEAVT
Mb2709c|M.bovis_AF2122/97 LAEQAEEA--EITLPSRNHAVVLVSKLHLPTLRALTYARATRPDVLEAVT
Rv2690c|M.tuberculosis_H37Rv LAEQAEEA--EITLPSRNHAVVLVSKLHLPTLRALTYARATRPDVLEAVT
MMAR_2024|M.marinum_M LEEQAAAHDNEVVLPSRNHALVLVSKLHLPTLRALAYARATRPDVLEAIT
MUL_3328|M.ulcerans_Agy99 LEEQAAAHGNEVVLPSRNHALVLVSKLHLPTLRALAYARATRPDVLEAIT
MAV_3581|M.avium_104 LAEQAADQD-DVVLPSRNHAVVLVSKLHLPTLRALAYARATRPDVLEAVT
* :.****.**:****: * ** ***:************:*
Mflv_3926|M.gilvum_PYR-GCK VSVDDAETRELVHKWENSDVSVPLKVIASPYREITRPVLDYVKRVTKDSP
Mvan_2474|M.vanbaalenii_PYR-1 VSVDDAETRQLVHKWEHSDISVPLKVVASPYREITRPVLDYVKRVGKDSP
MSMEG_2772|M.smegmatis_MC2_155 VSVDDAETRALVHEWEGSDVAVPLKVIASPYREITRPVLDYVKRITKESP
TH_2211|M.thermoresistible__bu VSVDDAETRELVHKWEESDVTVPLKVIASPYREVTRPVLDYVKRVTKESP
MAB_2993c|M.abscessus_ATCC_199 VSVDDRDTRELVREWETSDISTPLKVIASPYREITRPILDYVKRISKEAP
Mb2709c|M.bovis_AF2122/97 VNVDDAETRELVRQWQDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
Rv2690c|M.tuberculosis_H37Rv VNVDDAETRELVRQWQDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
MMAR_2024|M.marinum_M VSVDDVETRELVRQWEDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
MUL_3328|M.ulcerans_Agy99 VSVDDVETRELVRQWEDSDVSVPLKVIASPYREITRPVLDYVKRVSKESP
MAV_3581|M.avium_104 VSVDDAETRDLVHKWEEADISVPLKVIASPYREITRPVLEYVKRAREESP
*.*** :** **::*: :*::.****:******:***:*:**** :::*
Mflv_3926|M.gilvum_PYR-GCK RTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFEPNVMVTSVPWQLTS
Mvan_2474|M.vanbaalenii_PYR-1 RTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFEPNVMVTSVPWQLSS
MSMEG_2772|M.smegmatis_MC2_155 RTVVTVFIPEYVVGHWWEQILHNQSALRLKGRLLFMPNVMVTSVPWQLTS
TH_2211|M.thermoresistible__bu RTVVTVFIPEYVVGHWWEQLLHNQSALRLKGRLLFMPNVMVTSVPWQLSS
MAB_2993c|M.abscessus_ATCC_199 RTVVCIFIPEYVVGHWWEQVLHNQSALRLKGRLLFVPNVMVTSVPWQLDS
Mb2709c|M.bovis_AF2122/97 RTVVTVFIPEYVVGRWWEQLLHNQSALRLKGRLLFMPGVMVTSVPWQLTS
Rv2690c|M.tuberculosis_H37Rv RTVVTVFIPEYVVGRWWEQLLHNQSALRLKGRLLFMPGVMVTSVPWQLTS
MMAR_2024|M.marinum_M RTVVTVFIPEYVVGRWWEQLLHNQSAFRLKTRLLFMPGVMVTSVPWQLTS
MUL_3328|M.ulcerans_Agy99 RTVVTVFIPEYVVGRWWEQLLHNQSAFRLKTRLLFMPGVMVTSVPWQLTS
MAV_3581|M.avium_104 RTVVTVFIPEYVVGHWWEQVLHNQSALRLKGRLLFMPGVMVTSVPWQLSS
**** :********:****:******:*** **** *.********** *
Mflv_3926|M.gilvum_PYR-GCK SERLKKMAPQSAPGDARRGFLE
Mvan_2474|M.vanbaalenii_PYR-1 SERLKKLAPQSAPGDARRGFLD
MSMEG_2772|M.smegmatis_MC2_155 SERLKAIQRQSAPGDARRGFLE
TH_2211|M.thermoresistible__bu SERRRKLQPQWAPGDARRGFLE
MAB_2993c|M.abscessus_ATCC_199 SERRRELDRQSAPGDSRRGFDS
Mb2709c|M.bovis_AF2122/97 SERIKTLQPHAAPGDTRRGIFD
Rv2690c|M.tuberculosis_H37Rv SERIKTLQPHAAPGDT------
MMAR_2024|M.marinum_M SERINTLEPHAAPGDMRRGIFD
MUL_3328|M.ulcerans_Agy99 SERINTLEPHAAPGDTRRGIFD
MAV_3581|M.avium_104 SERRKTLEPHMAPGDARRGIFD
*** . : : ****