For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDNNAEFASRGRRSGEILAGRPPESGQPPRAPVHAPAIIFDHVTVAYGRGRKRSEALRNFNLRIARGETV ALLGPSGSGKSTALKALAGFVRPVFGTVRLDGEDVTDLPPAKRGIGVVVQSYALFPHMRVRDNVAFGLRA RRTKSDRIAECVDDALAMVSMSAYADRYPRELSGGQQQRVAIARALAIRPRVLLLDEPLAALDAQLRLSM IAELRQLQQSLPDTAMLYVTHDQSEALALADRIAVMRDAELVDVDSAHNLWTRPPTDFTANFLGGANLIP CTVGRVSGESALVSVGDKMLSVTVPTPEIGRDPWAPGASALICIRPHTLSIVGPTAPGALRARVANKVWR GASTRVSLAVSGLPDQLIDAEVPGHADVQPDSVVGVKFPEPAGALVAVPGEA
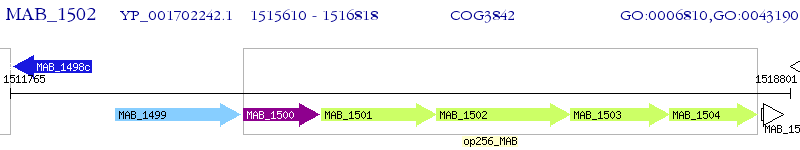
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1502 | - | - | 100% (402) | ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1655 | - | 1e-54 | 50.23% (219) | sulfate ABC transporter, ATP-binding protein SysA |
| M. abscessus ATCC 19977 | MAB_1375 | - | 4e-48 | 42.34% (248) | sugar ABC transporter, ATP-binding protein SugC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 3e-54 | 46.91% (243) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_1083 | - | 7e-63 | 43.96% (364) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 3e-54 | 46.91% (243) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01089 | - | 2e-48 | 37.29% (295) | putative ABC-transport protein, ATP-binding component |
| M. marinum M | MMAR_3715 | cysA1 | 4e-53 | 47.77% (224) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 1e-53 | 47.83% (230) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. smegmatis MC2 155 | MSMEG_0662 | - | 5e-63 | 43.67% (332) | putrescine transport ATP-binding protein PotG |
| M. thermoresistible (build 8) | TH_2424 | - | 4e-59 | 50.87% (230) | PUTATIVE sulfate ABC transporter, ATPase subunit |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 4e-53 | 47.77% (224) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_5739 | - | 1e-63 | 42.70% (363) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1083|M.gilvum_PYR-GCK ------MTNRG-------VG-----------------VQFDELTRVYGAT
Mvan_5739|M.vanbaalenii_PYR-1 ----MSVGTNG-------VG-----------------VQLDELTRVYGTT
MAB_1502|M.abscessus_ATCC_1997 MDNNAEFASRGRRSGEILAGRPPESGQPPRAPVHAPAIIFDHVTVAYGRG
Mb2419c|M.bovis_AF2122/97 ------MT---------------------------YAIVVADATKRYGDF
Rv2397c|M.tuberculosis_H37Rv ------MT---------------------------YAIVVADATKRYGDF
MMAR_3715|M.marinum_M MKKETSLTGPGT---------------------GGHAIVVRDAFKQYGDF
MUL_3658|M.ulcerans_Agy99 ------MTGPGT---------------------GGHAIVVRDAFKQYGDF
MAV_1785|M.avium_104 ------MTDAGTDR-------------------GDIAITVRDAYKRYGDF
TH_2424|M.thermoresistible__bu ------MT---------------------------DAITVRGANKRYGDF
MSMEG_0662|M.smegmatis_MC2_155 ------MTGG--------------------------AIRLHQLAKAFEGV
MLBr_01089|M.leprae_Br4923 ----------------------------------MAEIVLEHVNKNYPNG
: . :
Mflv_1083|M.gilvum_PYR-GCK R----ALDGLTLHIEPGELVALLGPSGGGKTTALRILAGLDEATSGTVSV
Mvan_5739|M.vanbaalenii_PYR-1 R----ALDGFTLHIEPGELVALLGPSGCGKTTALRILAGLDEATSGTVAV
MAB_1502|M.abscessus_ATCC_1997 RKRSEALRNFNLRIARGETVALLGPSGSGKSTALKALAGFVRPVFGTVRL
Mb2419c|M.bovis_AF2122/97 V----ALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTITI
Rv2397c|M.tuberculosis_H37Rv V----ALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTITI
MMAR_3715|M.marinum_M V----ALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDSGNITI
MUL_3658|M.ulcerans_Agy99 V----ALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDSGNITI
MAV_1785|M.avium_104 V----ALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTVTI
TH_2424|M.thermoresistible__bu A----ALDNVDFAVPAGSLTALLGPSGSGKSTLLRAIAGLDRPDTGTITI
MSMEG_0662|M.smegmatis_MC2_155 P----AVAGIDLDIPAGQFYSLLGASGCGKTTTLRMIAGFEKPDSGRIEL
MLBr_01089|M.leprae_Br4923 AT---AVHDLSITVADGEFLILIGPSGCGKTTTLNMIAGLEDISSGELRI
*: . : : *. *:*.** **:* *. :**: * : :
Mflv_1083|M.gilvum_PYR-GCK GGVDVGRVPANKRDMGMVFQAYSLFPHLTVLDNVAFGLKMRGKGKSDRLS
Mvan_5739|M.vanbaalenii_PYR-1 GGVDVSKVPANKRDMGMVFQAYSLFPHLTVLDNVAFGLKMRGKAKRDRLS
MAB_1502|M.abscessus_ATCC_1997 DGEDVTDLPPAKRGIGVVVQSYALFPHMRVRDNVAFGLRAR-RTKSDRIA
Mb2419c|M.bovis_AF2122/97 NGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAFGLKIRKRPKAE-IK
Rv2397c|M.tuberculosis_H37Rv NGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAFGLKIRKRPKAE-IK
MMAR_3715|M.marinum_M NGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAE-IR
MUL_3658|M.ulcerans_Agy99 NGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAE-IR
MAV_1785|M.avium_104 YGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKVRKRPKAE-IK
TH_2424|M.thermoresistible__bu NGRDVTDVPPQRRGIGFVFQHYAAFKHLTVRDNVAFGLKIRRRPKRE-IA
MSMEG_0662|M.smegmatis_MC2_155 DGRDVASDPPHKRPVNTVFQTYALFPFMSVWDNVAFGLRYQKAPRAE-VT
MLBr_01089|M.leprae_Br4923 DGDRVNEKAPKDRDIAMVFQSYALYPHMTVRQNIAFPLMLAKVKKAE-IA
* * .. * : *.* *: : .: * :*:*: * : : :
Mflv_1083|M.gilvum_PYR-GCK RAAD-MLDLVGLGELGGRYANELSGGQQQRVALARALAIRPRVLLLDEPL
Mvan_5739|M.vanbaalenii_PYR-1 RAAD-MLELVGLGELGGRYAKELSGGQQQRVALARALAIRPRVLLLDEPL
MAB_1502|M.abscessus_ATCC_1997 ECVDDALAMVSMSAYADRYPRELSGGQQQRVAIARALAIRPRVLLLDEPL
Mb2419c|M.bovis_AF2122/97 AKVDNLLQVVGLSGFQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEPF
Rv2397c|M.tuberculosis_H37Rv AKVDNLLQVVGLSGFQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEPF
MMAR_3715|M.marinum_M DKVDNLLEVVGLSGFHGRYPNQLSGGQRQRMALARALAVDPEVLLLDEPF
MUL_3658|M.ulcerans_Agy99 DKVDNLLEVVGLSGFHGRYPDQLSGGQRQRMALARALAVDPEVLLLDEPF
MAV_1785|M.avium_104 AKVDNLLEVVGLSGFQGRYPNQLSGGQRQRMALARALAVDPQVLLLDEPF
TH_2424|M.thermoresistible__bu EKVDNLLEVVGLAGFQNRYPNQLSGGQRQRMALARALAVDPEVLLLDEPF
MSMEG_0662|M.smegmatis_MC2_155 RRVGEALELVRMAEFAKRKPAQLSGGQQQRVALARALVLRPRVLLLDEPL
MLBr_01089|M.leprae_Br4923 QKVSETAQILDLTDLLDRKPSQLSGGQRQRVAMGRAIVRHPKAFLMDEPL
.. :: : * . :*****:**:*:.**:. *..:*:***:
Mflv_1083|M.gilvum_PYR-GCK SALDAKVRTQLRDEIRRVQLEVG-TTTLFVTHDQEEALAVADRVGVMNQG
Mvan_5739|M.vanbaalenii_PYR-1 SALDAKVRTQLRDEIRRVQLEVG-TTTLFVTHDQEEALAVADRVGVMNEG
MAB_1502|M.abscessus_ATCC_1997 AALDAQLRLSMIAELRQLQQSLPDTAMLYVTHDQSEALALADRIAVMRDA
Mb2419c|M.bovis_AF2122/97 GALDAKVREELRAWLRRLHDEVH-VTTVLVTHDQAEALDVADRIAVLHKG
Rv2397c|M.tuberculosis_H37Rv GALDAKVREELRAWLRRLHDEVH-VTTVLVTHDQAEALDVADRIAVLHKG
MMAR_3715|M.marinum_M GALDAKVREDLRAWLRRLHDEVH-VTTVLVTHDQAEALDVADRIAVLNSG
MUL_3658|M.ulcerans_Agy99 GALDAKVREDLRAWLRRLHDEVH-VTTVLVTHDQAEALDVADRIAVLNSG
MAV_1785|M.avium_104 GALDAKVREDLRAWLRRLHDEVH-VTTVLVTHDQAEALDVADRIAVLNQG
TH_2424|M.thermoresistible__bu GALDAKVREDLRTWLRRLHEEVH-VTTVLVTHDQSEALDVADRIAVLNNG
MSMEG_0662|M.smegmatis_MC2_155 GALDAKLRKQLQLELRALQREVG-ITFVYVTHDQEEALTMSDQIAVMAEG
MLBr_01089|M.leprae_Br4923 SNLDAKLRVRTRGEIARLQRRLG-ATTVYVTHDQTEAMTLGDRVVVMRSG
. ***::* : :: : : : ***** **: :.*:: *: ..
Mflv_1083|M.gilvum_PYR-GCK RLEQLAAPADLYAHPATPFVADFVG--LNNKVPASVS--G----GTATLL
Mvan_5739|M.vanbaalenii_PYR-1 RLEQLAAPADLYANPATPFVAEFVG--LNNKVPATVS--G----GSASLL
MAB_1502|M.abscessus_ATCC_1997 ELVDVDSAHNLWTRPPTDFTANFLG--GANLIPCTVGRVS----GESALV
Mb2419c|M.bovis_AF2122/97 RIEQVGSPTDVYDAPANAFVMSFLG--AVSTLNGSLVR------PHDIRV
Rv2397c|M.tuberculosis_H37Rv RIEQVGSPTDVYDAPANAFVMSFLG--AVSTLNGSLVR------PHDIRV
MMAR_3715|M.marinum_M RIEQVGSPTEVYDAPANAFVMSFLG--AVSALNGALVR------PHDIRV
MUL_3658|M.ulcerans_Agy99 RIEQVGSPTEVYDAPTNAFVMSFLG--AVSALNGALVR------PHDIRV
MAV_1785|M.avium_104 RIEQIGSPTEVYDAPTNAFVMSFLG--AVSTLNGTLVR------PHDIRV
TH_2424|M.thermoresistible__bu RIEQVGSPAEVYDSPANEFVMSFLG--AVSSLNGTLVR------PHDIWV
MSMEG_0662|M.smegmatis_MC2_155 RVEQVGAPTEIYSAPATTYVAGFLG--AANIFDAELTEAAGAT-GGTAVC
MLBr_01089|M.leprae_Br4923 VVQQIGTPDELYERPVNLFVAGFIGSPVMNFFPAKLTPNGLTLPLGELNL
: :: :. ::: * . :. *:* . . :
Mflv_1083|M.gilvum_PYR-GCK GVSVPALPGSVD--------------GDGVAMVRPEAVTVAADPAG----
Mvan_5739|M.vanbaalenii_PYR-1 GVSVAALPGSVD--------------GAGVAMVRPESVTVTADPAG----
MAB_1502|M.abscessus_ATCC_1997 SVGDKMLSVTVPTPEIGRD--PWAPGASALICIRPHTLSIVGPTAPGAL-
Mb2419c|M.bovis_AF2122/97 GRTPNMAVAAADGTAG----------STGVLRAVVDRVVVLGFEVRVEL-
Rv2397c|M.tuberculosis_H37Rv GRTPNMAVAAADGTAG----------STGVLRAVVDRVVVLGFEVRVEL-
MMAR_3715|M.marinum_M GRTPEMAVAAADGTGE----------STGVTRAVVDRIVVLGFEVRVEL-
MUL_3658|M.ulcerans_Agy99 GRTPEMAVAAADGTGE----------STGVTRAVVDRIVVLGFEVRVEL-
MAV_1785|M.avium_104 GRTPEMAVAAEDGTAE----------STGVARAIVDRVVKLGFEVRVEL-
TH_2424|M.thermoresistible__bu GRHPELPTS--DSTLQ----------ATGVVRARIDRVVMLGFEVRLEL-
MSMEG_0662|M.smegmatis_MC2_155 TAMSTRLSAWVDGAAAP---------GPAAIVIRPERITLQGPDEPVAAG
MLBr_01089|M.leprae_Br4923 SRDVQAMIGQHPVPDRVIVGVRPEHLTDAKLIDAHQHGTVLTFKVKVDLV
. .
Mflv_1083|M.gilvum_PYR-GCK ----TATVATVSFLGAITRVGITMPDGSLLSAQMSSTAARALTPGDPVTV
Mvan_5739|M.vanbaalenii_PYR-1 ----ASTVSSVSFLGAISRVHIMVPDGSTVSAQMASSAARGFAPGDPVTV
MAB_1502|M.abscessus_ATCC_1997 ----RARVANKVWRGASTRVSLAVSG---LPDQLIDAEVPGHADVQPDSV
Mb2419c|M.bovis_AF2122/97 ----TSAATGGAFTAQITRG-----DAEALALREGDTVYVRATRVPPIAG
Rv2397c|M.tuberculosis_H37Rv ----TSAATGGAFTAQITRG-----DAEALALREGDTVYVRATRVPPIAG
MMAR_3715|M.marinum_M ----TSAATGGAFTAQITRG-----DAEALALQEGDTVYVRATRVPPIAG
MUL_3658|M.ulcerans_Agy99 ----TSAATGGAFTAQITRG-----DAEALALQEGDTVYVRATRVPPIAG
MAV_1785|M.avium_104 ----TSAATGGPFTAQITRG-----DAEALALREGDTVYVRATRVPPITA
TH_2424|M.thermoresistible__bu ----TSATNNAPFTAQITRG-----DAEALGLRSGDTVYVRATRVP----
MSMEG_0662|M.smegmatis_MC2_155 SNVITGTVSHVVYLGASTQVHVDVGEANSLVVEVPNHAGPQSVSQQAGAR
MLBr_01089|M.leprae_Br4923 ES--LGADKYIYFTTTGCDVYSAQLDELASELEVRENQFVARVSAESKMA
. : . . .
Mflv_1083|M.gilvum_PYR-GCK G------VEPGGVLVTRPL-----------------
Mvan_5739|M.vanbaalenii_PYR-1 G------IEPRGVLVTRP------------------
MAB_1502|M.abscessus_ATCC_1997 VG--VKFPEPAGALVAVPGEA---------------
Mb2419c|M.bovis_AF2122/97 ---GVSGVDDAGVERVKVTST---------------
Rv2397c|M.tuberculosis_H37Rv ---GVSGVDDAGVERVKVTST---------------
MMAR_3715|M.marinum_M ---AAPAVPDDGAGSTPSAAQIGVGGQ---------
MUL_3658|M.ulcerans_Agy99 ---AAPAVPDDGAGSTPRAAQIGVGGQ---------
MAV_1785|M.avium_104 GATTVPAVSRDGADEATLTSA---------------
TH_2424|M.thermoresistible__bu ------ALPTGDVDERVAAR----------------
MSMEG_0662|M.smegmatis_MC2_155 VH-CVCTPDAVRVLTRSPVAVIADPVVEPETALSAH
MLBr_01089|M.leprae_Br4923 IGESIELAFGTAKIAVFDADSGVNLTVSAPTDA---