For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRRDTTSMSDVVAAARRALGAVGAFLPMHFTSMPSVGEQRAAVVRMEAAGYRTTWLNEPVGGKDVLVQAG LLLAATEHMTFATGIANIWARPAVTAHGGAAMLAQAYPDRFVLGLGAGYPSQAELVGSHFGSGLGAMRDY LSRMGETTPLPAVDVRYPRIVAANGPKMLALARDAADGAMPAGRPPAFTAQVRQALGRDKLIVVGIDVIV AGDGDDTKTIARNMVSMRLELPGVRGGLKSLGYADEEVADISDRLVDDLVAYGSPGDVATMVDRHLQAGA DHVVLMPANVETEVGVRYLEQLAPALLS
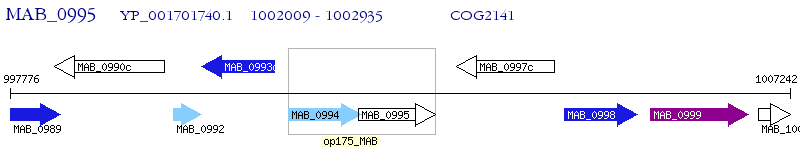
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0995 | - | - | 100% (308) | hypothetical protein MAB_0995 |
| M. abscessus ATCC 19977 | MAB_3778 | - | 2e-30 | 34.16% (243) | hypothetical protein MAB_3778 |
| M. abscessus ATCC 19977 | MAB_3245 | - | 2e-27 | 34.11% (258) | hypothetical protein MAB_3245 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3492 | - | 6e-32 | 36.25% (251) | hypothetical protein Mb3492 |
| M. gilvum PYR-GCK | Mflv_5006 | - | 1e-31 | 32.76% (290) | hypothetical protein Mflv_5006 |
| M. tuberculosis H37Rv | Rv3463 | - | 7e-32 | 36.25% (251) | hypothetical protein Rv3463 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1084 | - | 1e-31 | 35.61% (264) | hypothetical protein MMAR_1084 |
| M. avium 104 | MAV_4404 | - | 3e-31 | 35.68% (241) | hypothetical protein MAV_4404 |
| M. smegmatis MC2 155 | MSMEG_0741 | - | 2e-32 | 38.58% (254) | hypothetical protein MSMEG_0741 |
| M. thermoresistible (build 8) | TH_0027 | - | 5e-31 | 34.41% (247) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0842 | - | 1e-32 | 34.34% (297) | hypothetical protein MUL_0842 |
| M. vanbaalenii PYR-1 | Mvan_1367 | - | 2e-32 | 33.10% (290) | hypothetical protein Mvan_1367 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1084|M.marinum_M MPDPETDSDARSSASPVAGKPNLGRFGSFGRG---------VTPAQARDI
MUL_0842|M.ulcerans_Agy99 MPAPETDSDARSSASPAAGKPNLGRFGSFGRG---------VTPAQARDI
MAV_4404|M.avium_104 -------------MTQAAGKPDLGRFGSFGRG---------VTPQQAKDI
Mb3492|M.bovis_AF2122/97 --------MTNCAAGKPSSGPNLGRFGSFGRG---------VTPQQATEI
Rv3463|M.tuberculosis_H37Rv --------MTNCAAGKPSSGPNLGRFGSFGRG---------VTPQQATEI
Mflv_5006|M.gilvum_PYR-GCK ----------MTNSEGPSLKPALGRFGVWTGEP--------ATPDQAVEI
Mvan_1367|M.vanbaalenii_PYR-1 ----------MTASEGPSLKPALGRFGVWTGAP--------VSPDQAVEI
TH_0027|M.thermoresistible__bu ----------MT--DAVSLKPDLGRFGVWTGGP--------IPRELAAEI
MSMEG_0741|M.smegmatis_MC2_155 -----------MTEGQCARIDFPSRIGVWWASDTWSM---RDAQEVAREI
MAB_0995|M.abscessus_ATCC_1997 ----MRRDTTSMSDVVAAARRALGAVGAFLPMHFTSMPSVGEQRAAVVRM
: . .* : . :
MMAR_1084|M.marinum_M EALGYGAVWVGGSP-PAE-LAWVEPILAATTTLQVATGIVNIWSAPAKPV
MUL_0842|M.ulcerans_Agy99 EALGYGAVWVGGSP-PAE-LAWVEPILAATTTLQVATGIVNIWSAPAKPV
MAV_4404|M.avium_104 EALGYGAVWVGGSP-PAE-LAWVEPLLEATTTLQVATGIVNIWTAPAGPV
Mb3492|M.bovis_AF2122/97 EALGYGAVWVGGSP-PAA-LSWVEPILQATTTLCVATGIVNIWSAPAQRV
Rv3463|M.tuberculosis_H37Rv EALGYGAVWVGGSP-PAA-LSWVEPILQATTTLCVATGIVNIWSAPAQRV
Mflv_5006|M.gilvum_PYR-GCK EKLGYGTVWVGASP-AAD-LAFVEPILEKTETLQVATGIVNIWTADADEV
Mvan_1367|M.vanbaalenii_PYR-1 EKLGYGTVWVGASP-AAD-LAFVEPILEKTRSLQVATGIVNIWTADAAEV
TH_0027|M.thermoresistible__bu EKLGYRALWVGGSP-AAE-LDFVEPILEATETLQVATGIVNIWTAPARTV
MSMEG_0741|M.smegmatis_MC2_155 EALGFGSLFLPETV-AKECLTQSAAFLAATERLVIGTGIANIHVRLPSAA
MAB_0995|M.abscessus_ATCC_1997 EAAGYRTTWLNEPVGGKDVLVQAGLLLAATEHMTFATGIANIWARPAVTA
* *: : :: . * :* * : ..***.** . .
MMAR_1084|M.marinum_M AESFHRIEAAYPGRFLLGIGVGHPEAITEYHS--TSNAVDALIEYLNRLD
MUL_0842|M.ulcerans_Agy99 AESFHRIEAAYPGRFLLGIGVGHPEAITEYHS--TGNAVDALIEYLNRLD
MAV_4404|M.avium_104 AESFHRIEAAYPGRFLLGIGVGHPEAHTEYR-----KPYDALADYLTRLD
Mb3492|M.bovis_AF2122/97 AESFHRIEAAYPGRFLLGIGVGHAEMISEYR-----KPYNALVEYLDRLD
Rv3463|M.tuberculosis_H37Rv AESFHRIEAAYPGRFLLGIGVGHAEMISEYR-----KPYNALVEYLDRLD
Mflv_5006|M.gilvum_PYR-GCK AESYHRIEKAFPGRFLLGVGVGHPEHTQEYT-----KPYQALVDYLDKLD
Mvan_1367|M.vanbaalenii_PYR-1 SESYHRIEKAFPGRFLLGIGVGHPEHTQEYV-----KPYQALVDYLDKLD
TH_0027|M.thermoresistible__bu AQSFHRIEKAYPGRFVLGLGVGHPEATEEYA-----KPYEAMVTYLDVLD
MSMEG_0741|M.smegmatis_MC2_155 EAGARTLTALYPGRFVLGLGVSHGPLVERGMGGVYQKPLATMRTYLERMA
MAB_0995|M.abscessus_ATCC_1997 HGGAAMLAQAYPDRFVLGLGAGYPSQAELVGS-HFGSGLGAMRDYLSRMG
. : :*.**:**:*..: . :: ** :
MMAR_1084|M.marinum_M DYGVPANR------RVVAALGPRVLRLAARRSAGAHPYLTTPEHTAAARE
MUL_0842|M.ulcerans_Agy99 DYGVPANR------RVVAALGPRVLRLAARRSAGAHPYLTTPEHTGAARE
MAV_4404|M.avium_104 QYGVPAGR------RVVAALGPRVLRLSAERSAGAHPYLTTPEHTARARE
Mb3492|M.bovis_AF2122/97 DYGVPANR------RVVAALGPRVLGLSARRSAGAHPYLTTPEHTARARE
Rv3463|M.tuberculosis_H37Rv DYGVPANR------RVVAALGPRVLGLSARRSAGAHPYLTTPEHTARARE
Mflv_5006|M.gilvum_PYR-GCK AAKVPTSR------MVVAALGPKVLKLAAQRSAGAHPYLTTPVHTGQARE
Mvan_1367|M.vanbaalenii_PYR-1 AAKVPTSR------RVVAALGPKVLQLAAQRSAGAHPYLTTPVHTGQARE
TH_0027|M.thermoresistible__bu EATVPTSR------RVLAALGPRMLQLAAQRSAGAHPYLTTPQHTGQARN
MSMEG_0741|M.smegmatis_MC2_155 ALPEQIEPGAGRPPRLLAALGPKMIELSGTHADGAHPYLVLPEQTRTTRE
MAB_0995|M.abscessus_ATCC_1997 ETTPLPAVDVR-YPRIVAANGPKMLALARDAADGAMPAGRPPAFTAQVRQ
::** **::: *: : ** * * * .*:
MMAR_1084|M.marinum_M LIGPSAFLAPEHKVVLTTDAQRARAVGRKALDIYLNLANYLNSWKRLGFC
MUL_0842|M.ulcerans_Agy99 LIGPSAFLAPEHKVVLTTDAQRARAVGRKALDIYLNLANYLNSWKRLGFS
MAV_4404|M.avium_104 LIGPSAFLAPEHKVVLTTDAEHARAVGRKALDIYFNLANYRNNWKRLGFT
Mb3492|M.bovis_AF2122/97 LIGPSAFLAPEHKVVLTTDSARARTVGRQALDMYFNLANYRNNWKRLGFT
Rv3463|M.tuberculosis_H37Rv LIGPSAFLAPEHKVVLTTDSARARTVGRQALDMYFNLANYRNNWKRLGFT
Mflv_5006|M.gilvum_PYR-GCK LLGPTVLIAPEHKVVLTDDAEKAREIGRGTVDFYLGLSNYVNNWKRLGFT
Mvan_1367|M.vanbaalenii_PYR-1 LLGPTVLIAPEHKVVLTNDADKAREIGRGTVDFYLGLSNYLNNWKRLGFT
TH_0027|M.thermoresistible__bu RIGPSVFLAPEHKVVLDTDAAAAREIGRQTIGFYLRLRNYVNNWKQLGFT
MSMEG_0741|M.smegmatis_MC2_155 ILGEDKWIVSEQAVAVGGDDADQLRRAHQHLEVYSGLPNYRNSWLRQGFD
MAB_0995|M.abscessus_ATCC_1997 ALGRDKLIVVGIDVIVAGDGDDTKTIARNMVSMRLELPGVRGGLKSLGYA
:* :. * : * .: : . * . .. *:
MMAR_1084|M.marinum_M ERDIARPGSDRLVDAVVAYGTPAAIAARLNEHLQAGADHVPVQVLTKS--
MUL_0842|M.ulcerans_Agy99 ERDIARPGSDRLVDAVVAYGTPAAIAARLNEHLQAGADHVPVQVLTKS--
MAV_4404|M.avium_104 DDEITRPGSDRLVDALVAYGSVDRIAARLREHLDAGADHVPVQVLTKD--
Mb3492|M.bovis_AF2122/97 DDEVSRPGSDRLVDAVVAYGTPDAIAARLNEHLLAGADHVPIQVLTED--
Rv3463|M.tuberculosis_H37Rv DDEVSRPGSDRLVDAVVAYGTPDAIAARLNEHLLAGADHVPIQVLTED--
Mflv_5006|M.gilvum_PYR-GCK EEDLERPGSDRFIDAVVAYGSPDEIAARLSEHLAAGADHVAIQVLGGG--
Mvan_1367|M.vanbaalenii_PYR-1 DEDLTRPGSDRFIDAVVAHGSPDEIAARLTEHLAAGADHVAIQVLGGS--
TH_0027|M.thermoresistible__bu DDDIAEPGSDELIDAMVAHGTAEQVADALREHLDRGADHVAIQVLGGR--
MSMEG_0741|M.smegmatis_MC2_155 ESDLVRGGSDRLARAVVGMGSVDDAAKAVTAHLDAGADHVVVQVLGDTPT
MAB_0995|M.abscessus_ATCC_1997 DEEVAD-ISDRLVDDLVAYGSPGDVATMVDRHLQAGADHVVLMPANVET-
: :: **.: :*. *: * : ** ***** :
MMAR_1084|M.marinum_M ENLVPALAELA--------------------GPLGLTGA---------
MUL_0842|M.ulcerans_Agy99 ENLVPALAELA--------------------GPLGLAGA---------
MAV_4404|M.avium_104 ENLVSALAELA--------------------GPLGLKPS---------
Mb3492|M.bovis_AF2122/97 DNLVSALTELA--------------------KPLRLT-----------
Rv3463|M.tuberculosis_H37Rv DNLVSALTELA--------------------KPLRLT-----------
Mflv_5006|M.gilvum_PYR-GCK DELLSTLEELA--------------------GPLDLKAAD--------
Mvan_1367|M.vanbaalenii_PYR-1 DELLSTLEELA--------------------GPLGLTA----------
TH_0027|M.thermoresistible__bu ERLLPTLXXSPRCGPNWGRCTSWSRARVWWTSPRSPRSAGRPGSGSSM
MSMEG_0741|M.smegmatis_MC2_155 SDPRPVLRELA--------------------GAFGLK-----------
MAB_0995|M.abscessus_ATCC_1997 EVGVRYLEQLA---------------------PALLS-----------
. * . .