For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RELKVIGLSADGKRVICQDANGTDKFAIDADDRLRAAARGDLSRLGQIQIEMESSLRPREIQARIRAGAT VQQVATAAGVDVSRIERFAHPVLLERQRAAEMASAAHPVRNDGPAVSTLADTVAGAMFARGGDPDAIEWD AWKVEDGRWTVQLQWRTGRSDNKAHFRYTPGAHGGTVTALDDSARELIDPTFTRSLRPVAAVSPLLPEPE PELFTESPAPLPEPEPAVAEEHTETETTQAPAPRRSRRSKPAVPTWEDVLLGVRSSGQN*
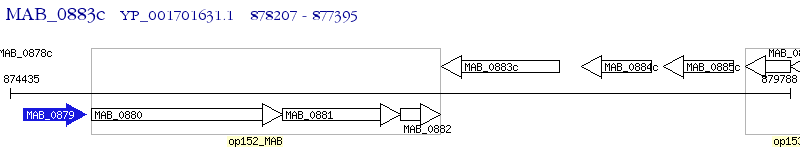
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0883c | - | - | 100% (270) | hypothetical protein MAB_0883c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0907c | - | 3e-84 | 58.30% (271) | hypothetical protein Mb0907c |
| M. gilvum PYR-GCK | Mflv_1717 | - | 1e-91 | 62.04% (274) | hypothetical protein Mflv_1717 |
| M. tuberculosis H37Rv | Rv0883c | - | 3e-84 | 58.30% (271) | hypothetical protein Rv0883c |
| M. leprae Br4923 | MLBr_02137 | - | 3e-79 | 56.46% (271) | hypothetical protein MLBr_02137 |
| M. marinum M | MMAR_4649 | - | 7e-84 | 56.46% (271) | hypothetical protein MMAR_4649 |
| M. avium 104 | MAV_1010 | - | 7e-77 | 55.15% (272) | hypothetical protein MAV_1010 |
| M. smegmatis MC2 155 | MSMEG_5685 | - | 6e-91 | 61.31% (274) | DNA-binding protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0267 | - | 2e-84 | 56.83% (271) | hypothetical protein MUL_0267 |
| M. vanbaalenii PYR-1 | Mvan_5034 | - | 4e-90 | 61.03% (272) | hypothetical protein Mvan_5034 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02137|M.leprae_Br4923 MRELKVVGLDPDSKTILCEGDIPGERFKLPADDRLRAAVRGDTTLLDQPQ
MAV_1010|M.avium_104 MRELKVVGLDADSKYLICESDDPAEQFKLPADDHLRAVLRHEAEPPEQPQ
MMAR_4649|M.marinum_M MRELRVVGLDADGKYILCEGVDSSEKYKLPADDRLRAALKRQSIPPEQPQ
MUL_0267|M.ulcerans_Agy99 MRELRVVGLDADGKYILCEGVDSTEKYKLPADDRLRAALKRQPIPPEQPQ
Mb0907c|M.bovis_AF2122/97 MRELKVVGLDADGKNIICQGAIPSEQFKLPVDDRLRAALRDDSVQPEQAQ
Rv0883c|M.tuberculosis_H37Rv MRELKVVGLDADGKNIICQGAIPSEQFKLPVDDRLRAALRDDSVQPEQAQ
Mflv_1717|M.gilvum_PYR-GCK MRELKVVGLDVDGTRIICETGDSGEKFVLRADDRLKAAVRGDRAASNQTT
Mvan_5034|M.vanbaalenii_PYR-1 MRELRVVGLDVDGRRIICETGDSGEKFVLRSDDRLKAAVRGDRAGSNQTT
MSMEG_5685|M.smegmatis_MC2_155 MRELRVVGLDVDGKRIICETADSAEMFTLRVDDRLRAALRGDKVASNQTE
MAB_0883c|M.abscessus_ATCC_199 MRELKVIGLSADGKRVICQDANGTDKFAIDADDRLRAAARGDLSRLGQIQ
****:*:**. *. ::*: : : : **:*:*. : : *
MLBr_02137|M.leprae_Br4923 LDIQVTNMLSPKEIQARIRAGASVEQVAMASGSDIARIRRFAHPVLLERS
MAV_1010|M.avium_104 LEIEVTNMLSPKEIQAKIRAGASVEQVAAASGSDVSRVRRFAHPVLLERY
MMAR_4649|M.marinum_M LDIEVTNMLSPKEIQARIRAGASVEQVATASGSDIARIQRFAHPVLLERS
MUL_0267|M.ulcerans_Agy99 LDIEVTNMLSPKEIQARIRAGASVEQVATASGSDIARIQRFAHPVLLERS
Mb0907c|M.bovis_AF2122/97 LDIEVTNVLSPKEIQARIRAGASVEQVAAASGSDIARIRRFAHPVLLERS
Rv0883c|M.tuberculosis_H37Rv LDIEVTNVLSPKEIQARIRAGASVEQVAAASGSDIARIRRFAHPVLLERS
Mflv_1717|M.gilvum_PYR-GCK IDVEVPNMLRPREIQSKIRAGASVEQIASAAGVDIARVERFAHPVLLERS
Mvan_5034|M.vanbaalenii_PYR-1 IDVEVPTLLRPREIQSKIRAGASVEQVAASAGVDIARVERFAHPVLLERA
MSMEG_5685|M.smegmatis_MC2_155 IDLEVQSVLRPKEIQAKIRAGASVEQLAAAAGVPVERVERFAHPVLLERA
MAB_0883c|M.abscessus_ATCC_199 IEMESS--LRPREIQARIRAGATVQQVATAAGVDVSRIERFAHPVLLERQ
:::: * *:***::*****:*:*:* ::* : *:.**********
MLBr_02137|M.leprae_Br4923 RAAELATAAHPVLADGPAVLTLLETVSAALVKRGLEPDKLSWDAWRNEDS
MAV_1010|M.avium_104 RAAELATAAHPMLADGPAVLTLLETVTAAFVTRGLRHDKLSWDAWRNEDN
MMAR_4649|M.marinum_M RAAELATAAHPMLADGPAVLTLLETITAALVTRGLNPERLDWDAWRNEDG
MUL_0267|M.ulcerans_Agy99 RAAELATAAHPMLADGPAVLTLLETITAALVTRGLNPERLDWDAWRNEDG
Mb0907c|M.bovis_AF2122/97 RAAELATAAHPVLADGPAVLTMQETVAAALVARGLNPDSLTWDAWRNEDS
Rv0883c|M.tuberculosis_H37Rv RAAELATAAHPVLADGPAVLTMQETVAAALVARGLNPDSLTWDAWRNEDS
Mflv_1717|M.gilvum_PYR-GCK RAAELATAAHPVLADGPSVLTLLETVSTALIARGLDPEATTWDAWRNEDS
Mvan_5034|M.vanbaalenii_PYR-1 RAAELASAAHPVLADGPSVLTLLETVTSALIARGLDPDGANWDAWRNEDS
MSMEG_5685|M.smegmatis_MC2_155 RAAELATAAHPVLADGPSVLTLLETVTKALVARGLDPDAVAWDAWRNEDS
MAB_0883c|M.abscessus_ATCC_199 RAAEMASAAHPVRNDGPAVSTLADTVAGAMFARGGDPDAIEWDAWKVEDG
****:*:****: ***:* *: :*:: *:. ** : ****: **.
MLBr_02137|M.leprae_Br4923 RWTVQLMWHAGRSDNLAHFCFTPGAHGGTVTASDDAANELIDPDFNRPLR
MAV_1010|M.avium_104 RWTVQLAWKVGRSDNVAHFCFTPGAHGGTVTAIDDAATALIDPNFELPLR
MMAR_4649|M.marinum_M RWTVQLAWKAGRSDNLAHFRFAPGAHGGTVTAIDDAASELIDPEYKAPLR
MUL_0267|M.ulcerans_Agy99 RWTVQLAWKAGRSDNLAHFRFAPGAHGGTVTAIDDAASELIDPEYKAPLR
Mb0907c|M.bovis_AF2122/97 RWTVQLAWKAGRSDNLAHFRFTPGAHGGTATAIDDTAHELINPTFNRPLR
Rv0883c|M.tuberculosis_H37Rv RWTVQLAWKAGRSDNLAHFRFTPGAHGGTATAIDDTAHELINPTFNRPLR
Mflv_1717|M.gilvum_PYR-GCK RWTVQLAWKAGLSDNVAHFRYAPGAHGGTVTAFDDAASELIDPTFARPLR
Mvan_5034|M.vanbaalenii_PYR-1 RWTVQLAWKAGLSDNVAHFRYAPGAHGGTVTAFDDAAAELIDPNFARPLR
MSMEG_5685|M.smegmatis_MC2_155 RWTVQLAWKAGRSDNVAHFRFTPGSHGGLVTASDDAAHQLINPDFARPLR
MAB_0883c|M.abscessus_ATCC_199 RWTVQLQWRTGRSDNKAHFRYTPGAHGGTVTALDDSARELIDPTFTRSLR
****** *:.* *** *** ::**:*** .** **:* **:* : .**
MLBr_02137|M.leprae_Br4923 PLARVAHLDFVEPATPAT------------VTPDNQLVS-----------
MAV_1010|M.avium_104 PLARVAHVDFDEPAKPPTAEAPA----AEPAEPEEKPVH-----------
MMAR_4649|M.marinum_M PLAPVSHLAFDEPAKPAKQAKPAKPAKQTPPAPAEQPAS-----------
MUL_0267|M.ulcerans_Agy99 PLAPVSHLAFDEPAKPAKQAKPAKPPKQTPPAPAEQPAS-----------
Mb0907c|M.bovis_AF2122/97 PLAPVAHLDFDEPEPAQP----------TLTVPSAQPVS-----------
Rv0883c|M.tuberculosis_H37Rv PLAPVAHLDFDEPEPAQP----------TLTVPSAQPVS-----------
Mflv_1717|M.gilvum_PYR-GCK PLAPVAQLALGEPEPAPAPTP------APAPAPAPAPESIPEPQAEAPED
Mvan_5034|M.vanbaalenii_PYR-1 PVAAVAQLALGEPEDPHAPVQ------AATPAPEPAPVETPEP-------
MSMEG_5685|M.smegmatis_MC2_155 PVAAVPQLEFEAPQPAQPAAPPQPAQAVPTPEPAPAPEQEPEP-------
MAB_0883c|M.abscessus_ATCC_199 PVAAVSPL-LPEPEPELFTESP-----APLPEPEPAVAEEHTET----ET
*:* *. : : * *
MLBr_02137|M.leprae_Br4923 --------SRRGKPTIPAWEDVLLGVRSAG-QR
MAV_1010|M.avium_104 --------TRRGKPVIPAWEDVLLGVRSGG-QR
MMAR_4649|M.marinum_M --------SRRGKPAIPAWEDVLLGVRSSG-QR
MUL_0267|M.ulcerans_Agy99 --------SRRGKPAIPAWEDVLLGVRSSG-QR
Mb0907c|M.bovis_AF2122/97 --------NRRGKPAIPAWEDVLLGVRSGG-RR
Rv0883c|M.tuberculosis_H37Rv --------NRRGKPAIPAWEDVLLGVRSGG-RR
Mflv_1717|M.gilvum_PYR-GCK TPAAKTSRPRKSRPAVPAWEDVLLGVRSSGGQR
Mvan_5034|M.vanbaalenii_PYR-1 APTVKTPRPRKSRPAVPAWEDVLLGVRSSGGQR
MSMEG_5685|M.smegmatis_MC2_155 APAPAPKRSRKARATVPAWEDVLLGVRSSGTQR
MAB_0883c|M.abscessus_ATCC_199 TQAPAPRRSRRSKPAVPTWEDVLLGVRSSG-QN
*:.:..:*:**********.* :.