For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTATIQETKAHAVATSGSIAHLDFVDQASFLGLRATEMATLSQMVWIYERDIDTDRLRRFHANLGRGLFG RRIQRSRLPFGRHRWVLDPACHPIDVELAPLPRAELQDWVHRHAQLPIDPEHGPSWRLGVTRFTDGAAAV SLVVSHSIADGVGLCRGIADAVNDAELPLGYAEPQARKRFRSMREDLRRARRDLPDTARAFRKATKLLTA RRQDIKHAGLPTIAAHVDRVAAATVYLDENHWDTHAHGRGGTSNSLVGAFAAQLAAKAGRLGRDGRVTLS MPVNERTEGDHRANAMTSVMFAIDPVRLTTDLSGIRSATKKVLSGLRDAPDEKWALLPLTPFIPKAAARR LADVALEYNDYPVGCSNVGALPAEVNRPDGTEADYIFCKLAEQRVRPDKVLSTYGQLFLASGRANRKVFL AVVAYQPGVIESRADLTTLLAETLDDFGLHAVIE
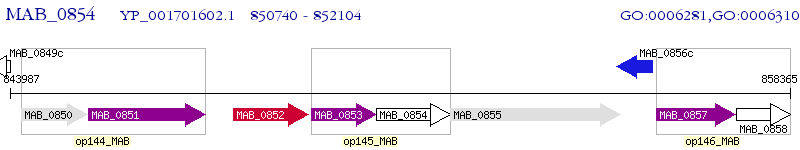
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0854 | - | - | 100% (454) | hypothetical protein MAB_0854 |
| M. abscessus ATCC 19977 | MAB_4701 | - | 6e-86 | 40.09% (439) | hypothetical protein MAB_4701 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0439 | - | 6e-05 | 26.20% (271) | hypothetical protein Mflv_0439 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2354 | - | 7e-93 | 42.70% (452) | hypothetical protein MMAR_2354 |
| M. avium 104 | MAV_4881 | - | 7e-91 | 44.14% (435) | hypothetical protein MAV_4881 |
| M. smegmatis MC2 155 | MSMEG_4729 | - | 6e-95 | 43.05% (453) | hypothetical protein MSMEG_4729 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1530 | - | 3e-92 | 42.70% (452) | hypothetical protein MUL_1530 |
| M. vanbaalenii PYR-1 | Mvan_2253 | - | 7e-05 | 27.69% (242) | hypothetical protein Mvan_2253 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2354|M.marinum_M --MLNESRTRSSSAMTDN---RIAFLDQAAFLRLRATGFETVGQTLWIYD
MUL_1530|M.ulcerans_Agy99 --MLNESRTRSSAAMTDN---RIAFLDQAAFLRLRATGFETVGQTLWIYD
MAV_4881|M.avium_104 ---------------MSN---VLDLADQTLFLGERATGATSLLQCVWVYD
MAB_0854|M.abscessus_ATCC_1997 -MTATIQETKAHAVATSGSIAHLDFVDQASFLGLRATEMATLSQMVWIYE
MSMEG_4729|M.smegmatis_MC2_155 MRFGDSSMTPSTTPATVATDDRLSHTDQLLFLGQRATGQELVAQCVWVYE
Mflv_0439|M.gilvum_PYR-GCK ------------MKRLRGWDAVLLYSETPSVHMHTLKLAVIELDDLGGAK
Mvan_2253|M.vanbaalenii_PYR-1 ------------MRRLNGVDAMMLYSETPEIHMHTLKIGVLDVSDVEGYD
: : . . . : .
MMAR_2354|M.marinum_M READMDGLRRFNENLGYGLLGRRIERSPLPFGRPRWVAYH-EAPDIDVAA
MUL_1530|M.ulcerans_Agy99 READMDGLRRSNENLGYGLLGRRIERSPLPFGRPRWVAYH-EAPDIDVAA
MAV_4881|M.avium_104 RAIDLAGLRRFHRHLGAGRLARRIECSPLPFGRHRWVSTG-AQGALEVVA
MAB_0854|M.abscessus_ATCC_1997 RDIDTDRLRRFHANLGRGLFGRRIQRSRLPFGRHRWVLDP-ACHPIDVEL
MSMEG_4729|M.smegmatis_MC2_155 RPVEIDKLRDFHRNFGHGLAGRLIERSPLPFGRHRWVAAPGPAAPLDFAE
Mflv_0439|M.gilvum_PYR-GCK FGVEELRKVIHGRLYKLDPFRYELVDIPFKFHHPMWRENAEVDLDYHVRS
Mvan_2253|M.vanbaalenii_PYR-1 F--DLFRRAALPRLHALAPLRYQLVDIPLKFHHPMWVQNAEIDVDYHLRR
: : : * : * ..
MMAR_2354|M.marinum_M T----PRPRSEITAWLDERAQIPVDPEYGPCWHLGVLP-LQDGGTAVSVV
MUL_1530|M.ulcerans_Agy99 T----PRPRSEITAWLDERAQIPVDPEYGPCWHLGVLP-LQDGGTAVSVV
MAV_4881|M.avium_104 V----PRPRDAVDAWLTEQAGRRPDAEHGPGWHLAVLP-LTDGGAVVSLV
MAB_0854|M.abscessus_ATCC_1997 A----PLPRAELQDWVHRHAQLPIDPEHGPSWRLGVTR-FTDGAAAVSLV
MSMEG_4729|M.smegmatis_MC2_155 Q----PRPREELGDWADARAQLPIDPEVGPGWHMGVLS-MTDGSTAVSLV
Mflv_0439|M.gilvum_PYR-GCK VRVDAPGGRRELDEAVGRIASTPLDRSR-PLWEMYLIEGLADGRIAVLGK
Mvan_2253|M.vanbaalenii_PYR-1 ARVPAPGGRRELDQLIGEIASTPLDRDH-PLWEMYIAEGLADNKIAVIHK
* * : * * . * *.: : : *. .*
MMAR_2354|M.marinum_M ITHSVVDGVAGGLAVFEAINGIT----------RDFGYPPPGSRGRMEGL
MUL_1530|M.ulcerans_Agy99 ITHSVVDGVAGGLAVFEAINGIT----------RDFGYPPPGSRGRMEGL
MAV_4881|M.avium_104 TTHCLTDGVGLCEALADAARGRA----------DTVRWPAAAARPRWSAL
MAB_0854|M.abscessus_ATCC_1997 VSHSIADGVGLCRGIADAVNDAE----------LPLGYAEPQARKRFRSM
MSMEG_4729|M.smegmatis_MC2_155 ISHCLIDGLGTLQTIADAVKGET----------VDFGYAAPGSRGRLQAV
Mflv_0439|M.gilvum_PYR-GCK IHHALADGVASANLLARGMDLQDGPQVD---RDSYATDPAPSKGELVRTA
Mvan_2253|M.vanbaalenii_PYR-1 VHHVLADGVASANQLAMAIQPRE-PEVGGQLAESSADSRAPRN--LLKAA
* : **:. : . .
MMAR_2354|M.marinum_M LVDLWDALRGLPEVFRALLALILIVLRRDTGAAGQTGKPPTVSD-----G
MUL_1530|M.ulcerans_Agy99 LVDLWDALRGLPEVFRALLALILIVLRRDTGAAGQTGKPPTVSD-----G
MAV_4881|M.avium_104 AEDARQTARDTGDVGRAVRAAARMLRQGRGGAALARGR-ATRRR-----G
MAB_0854|M.abscessus_ATCC_1997 REDLRRARRDLPDTARAFRKATKLLTARR--QDIKHAGLPTIAA-----H
MSMEG_4729|M.smegmatis_MC2_155 LADLRQTARDLPQTAATLVRAGRLAIRRRHEVALSAKPRRDFGP-----D
Mflv_0439|M.gilvum_PYR-GCK FTDHLRQIRRFPDVVRYTAAGMRRVRSSGQKLS--PELTRPFTPPPSFMN
Mvan_2253|M.vanbaalenii_PYR-1 ARDHLRQLRRLPRLVNDTAAGVSRVRRLARERGGHPDLARNFAPPPTFLN
* *
MMAR_2354|M.marinum_M ADEPIVTPSVTAFCDLETWDSCALDLSGSSGSLFVSFAT-RLAQLMGRVR
MUL_1530|M.ulcerans_Agy99 ADEPIVTPSVTAFCDLETWDSCALDLSGSSGSLFVSFAT-RLAQLMGRVR
MAV_4881|M.avium_104 TQDRVTVPAQTIFVDADEWDARARALGGTSNALLAGLAA-RLAQRVGRV-
MAB_0854|M.abscessus_ATCC_1997 VDR---VAAATVYLDENHWDTHAHGRGGTSNSLVGAFAA-QLAAKAGRL-
MSMEG_4729|M.smegmatis_MC2_155 GRSDVVAPAIACYVPLEEWDLRAAALGGNSHSLVAAFAA-KLGELMGRRR
Mflv_0439|M.gilvum_PYR-GCK HQIDATRKFATATLALDDVKQTGKHLGATINDMVLAMSAGTLRKLLLRYD
Mvan_2253|M.vanbaalenii_PYR-1 HVVTPGRRFATAPLALADVKETARHLGVTLNDVVLATAAGALRELQLRYD
: . . . . :. . :: * *
MMAR_2354|M.marinum_M -ATDGAVTIIMPVSERTNDDTRANAVTAITFDLDPDRARTDLQFIRNETK
MUL_1530|M.ulcerans_Agy99 -VTDGAVTIIMPVSERTNDDTRANAVTAITFDLDPDRARTDLQFIRNETK
MAV_4881|M.avium_104 -ARDGSVTVGMPVNERVAGDTRANAVTNVDVLLGPAAIGTDLRAIRAAVK
MAB_0854|M.abscessus_ATCC_1997 -GRDGRVTLSMPVNERTEGDHRANAMTSVMFAIDPVRLTTDLSGIRSATK
MSMEG_4729|M.smegmatis_MC2_155 -SDDGTVLLHVPISDRAPGDTRGNAAYIVNAHLDPTDVTKDLSGARSAIR
Mflv_0439|M.gilvum_PYR-GCK GQADHPLLASVPVSFDFSRDRISGNYFTGVLVSIPVELDDPVERVRAAHD
Mvan_2253|M.vanbaalenii_PYR-1 QKADAPLIAGVPVSFSTSPDRLVGNEFTYMTPSLPVHVADPMARVRLTAT
* : :*:. * . * : *
MMAR_2354|M.marinum_M QSLSALKDTPNELLKPLP-----LVPYTPKWLARKMEVLAMGTSDLPVGC
MUL_1530|M.ulcerans_Agy99 QSLSALKDTPNELLKPLP-----LVPYTPKWLARKMEVLAMGTSDLPVGC
MAV_4881|M.avium_104 HALVRHHDLPDERWALLP-----LVPLIPKRVFRRLLGVVTG-GATTVVS
MAB_0854|M.abscessus_ATCC_1997 KVLSGLRDAPDEKWALLP-----LTPFIPKAAARRLADVALEYNDYPVGC
MSMEG_4729|M.smegmatis_MC2_155 QALEAYRQEPDETLQLLP-----LTPFIPQRAVARGSQVVLNLTDLPVSC
Mflv_0439|M.gilvum_PYR-GCK AAVAGKESNNLLGPELVSQWSAYLPPAPAEALFRWLGTKDGQNKVMNLPI
Mvan_2253|M.vanbaalenii_PYR-1 ATAIAKENHRLLGPTLLPSWLAYLPPATTPRIFRMQARRLESAMVMNLTV
.. :. * * . :
MMAR_2354|M.marinum_M SNLGELSQALTRVDGTGARYSSARMFNRGATRQRFERESGELYLFSGRLE
MUL_1530|M.ulcerans_Agy99 SNLGELSQALTRVDGTGARYSSARMFNRGATRQRFERESGELYLFSGRLE
MAV_4881|M.avium_104 SNLGALDPAANRPDGTDADYFAMRSVYPGVTDATMQRTNGALALLSGRVH
MAB_0854|M.abscessus_ATCC_1997 SNVGALPAEVNRPDGTEADYIFCKLAEQRVRPDKVLSTYGQLFLASGRAN
MSMEG_4729|M.smegmatis_MC2_155 SNLGDIDPLVSRIDGAAAEYLILRGVDRRVSREFLENRNGLLTVLAGRVD
Mflv_0439|M.gilvum_PYR-GCK SNVPGPRERARVGGALVTEIYSVGPLTVGSGINITVWSYVDQLNISVLTD
Mvan_2253|M.vanbaalenii_PYR-1 SNVAGPKRRGVIEGAVVDEIYSVGPIVAGSGMNITVWSYVDQLSISVLTD
**: .. : .
MMAR_2354|M.marinum_M NSVFISVSAYQPGAENSNQYLRGLVEQTLADYSLKAELLG---------
MUL_1530|M.ulcerans_Agy99 NSVFISVSAYQPGAENSNQYLRGLVEQTLADYSLKAELLG---------
MAV_4881|M.avium_104 DRVFVSVLGYELGRQNSNEALRQALSGALNDFSLTGATGWNPVRAVATR
MAB_0854|M.abscessus_ATCC_1997 RKVFLAVVAYQPGVIESRADLTTLLAETLDDFGLHAVIE----------
MSMEG_4729|M.smegmatis_MC2_155 DKMSISVIGYRPGGPNDKPQLRELVAKALAEFDLVGTIA----------
Mflv_0439|M.gilvum_PYR-GCK GKTLEDPHELTSAMLDDFVEIRRAAGLSEELTVVETALPN---------
Mvan_2253|M.vanbaalenii_PYR-1 DRTLKDPHEATDALIASFAEIRRAAGLPEELTPVEKALPVAPAL-----
. . : . :