For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRFGDSSMTPSTTPATVATDDRLSHTDQLLFLGQRATGQELVAQCVWVYERPVEIDKLRDFHRNFGHGLA GRLIERSPLPFGRHRWVAAPGPAAPLDFAEQPRPREELGDWADARAQLPIDPEVGPGWHMGVLSMTDGST AVSLVISHCLIDGLGTLQTIADAVKGETVDFGYAAPGSRGRLQAVLADLRQTARDLPQTAATLVRAGRLA IRRRHEVALSAKPRRDFGPDGRSDVVAPAIACYVPLEEWDLRAAALGGNSHSLVAAFAAKLGELMGRRRS DDGTVLLHVPISDRAPGDTRGNAAYIVNAHLDPTDVTKDLSGARSAIRQALEAYRQEPDETLQLLPLTPF IPQRAVARGSQVVLNLTDLPVSCSNLGDIDPLVSRIDGAAAEYLILRGVDRRVSREFLENRNGLLTVLAG RVDDKMSISVIGYRPGGPNDKPQLRELVAKALAEFDLVGTIA
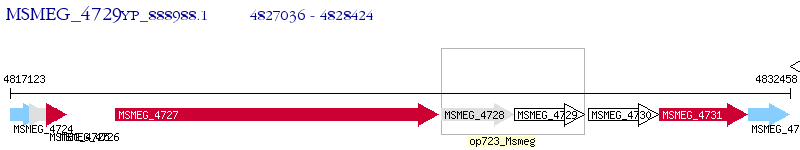
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4729 | - | - | 100% (462) | hypothetical protein MSMEG_4729 |
| M. smegmatis MC2 155 | MSMEG_4730 | - | e-113 | 48.21% (446) | hypothetical protein MSMEG_4730 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4701 | - | 1e-102 | 44.62% (455) | hypothetical protein MAB_4701 |
| M. marinum M | MMAR_2354 | - | 4e-99 | 42.06% (447) | hypothetical protein MMAR_2354 |
| M. avium 104 | MAV_4881 | - | 9e-89 | 42.01% (438) | hypothetical protein MAV_4881 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1530 | - | 7e-98 | 41.83% (447) | hypothetical protein MUL_1530 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2354|M.marinum_M ----------MLNESRTRSSSAMTDNRIAFLDQAAFLRLRATGFETVGQT
MUL_1530|M.ulcerans_Agy99 ----------MLNESRTRSSAAMTDNRIAFLDQAAFLRLRATGFETVGQT
MAV_4881|M.avium_104 -----------------------MSNVLDLADQTLFLGERATGATSLLQC
MSMEG_4729|M.smegmatis_MC2_155 ----MRFGDSSMTP-STTPATVATDDRLSHTDQLLFLGQRATGQELVAQC
MAB_4701|M.abscessus_ATCC_1997 MPGQIRHMRKSLSRRRTSKGAAAMDNRLALADEALLAEHHAAGMNVIIQV
.: : *: : :*:* : *
MMAR_2354|M.marinum_M LWIYDREADMDGLRRFNENLGYGLLGRRIERSPLPFGRPRWVAYHEAPD-
MUL_1530|M.ulcerans_Agy99 LWIYDREADMDGLRRSNENLGYGLLGRRIERSPLPFGRPRWVAYHEAPD-
MAV_4881|M.avium_104 VWVYDRAIDLAGLRRFHRHLGAGRLARRIECSPLPFGRHRWVSTGAQGA-
MSMEG_4729|M.smegmatis_MC2_155 VWVYERPVEIDKLRDFHRNFGHGLAGRLIERSPLPFGRHRWVAAPGPAAP
MAB_4701|M.abscessus_ATCC_1997 TWLYSHAIDLDALQRFRHNLGRGLLGRRIEAPALPIARHRWVIDHGPAD-
*:*.: :: *: ..::* * .* ** ..**:.* ***
MMAR_2354|M.marinum_M IDVAATPRPRSEITAWLDERAQIPVDPEYGPCWHLGVLPLQDGGTAVSVV
MUL_1530|M.ulcerans_Agy99 IDVAATPRPRSEITAWLDERAQIPVDPEYGPCWHLGVLPLQDGGTAVSVV
MAV_4881|M.avium_104 LEVVAVPRPRDAVDAWLTEQAGRRPDAEHGPGWHLAVLPLTDGGAVVSLV
MSMEG_4729|M.smegmatis_MC2_155 LDFAEQPRPREELGDWADARAQLPIDPEVGPGWHMGVLSMTDGSTAVSLV
MAB_4701|M.abscessus_ATCC_1997 IDIEERARPRAEFSEWVDERSQLHIDPESGPGWHIGVLPLQDGTTAISLV
::. .*** . * :: *.* ** **:.**.: ** :.:*:*
MMAR_2354|M.marinum_M ITHSVVDGVAGGLAVFEAINGITRDFGYPPPGSRGRMEGLLVDLWDALRG
MUL_1530|M.ulcerans_Agy99 ITHSVVDGVAGGLAVFEAINGITRDFGYPPPGSRGRMEGLLVDLWDALRG
MAV_4881|M.avium_104 TTHCLTDGVGLCEALADAARGRADTVRWPAAAARPRWSALAEDARQTARD
MSMEG_4729|M.smegmatis_MC2_155 ISHCLIDGLGTLQTIADAVKGETVDFGYAAPGSRGRLQAVLADLRQTARD
MAB_4701|M.abscessus_ATCC_1997 LSHNLIDGLGLALVIVEAVMGRTRDLGYPLPLSRRRLRALGQDVRQTARD
:* : **:. .: :* * : . :. . :* * .: * :: *.
MMAR_2354|M.marinum_M LPEVFRALLALILIVLRRDTGAAGQ-----TGKPPTVSDGADEPIVTPSV
MUL_1530|M.ulcerans_Agy99 LPEVFRALLALILIVLRRDTGAAGQ-----TGKPPTVSDGADEPIVTPSV
MAV_4881|M.avium_104 TGDVGRAVRAAARMLRQGRGGAALA-----RGR-ATRRRGTQDRVTVPAQ
MSMEG_4729|M.smegmatis_MC2_155 LPQTAATLVRAGRLAIRRRHEVALS---AKPRRDFGPDGRSD--VVAPAI
MAB_4701|M.abscessus_ATCC_1997 VPQATRALVAAVRLGRKQARDKNLPGLAPVAVRPAGSTAPSDEVVLVPAI
:. :: : : : :: : .*:
MMAR_2354|M.marinum_M TAFCDLETWDSCALDLSGSSGSLFVSFATRLAQLMGRVRATDGAVTIIMP
MUL_1530|M.ulcerans_Agy99 TAFCDLETWDSCALDLSGSSGSLFVSFATRLAQLMGRVRVTDGAVTIIMP
MAV_4881|M.avium_104 TIFVDADEWDARARALGGTSNALLAGLAARLAQRVGRV-ARDGSVTVGMP
MSMEG_4729|M.smegmatis_MC2_155 ACYVPLEEWDLRAAALGGNSHSLVAAFAAKLGELMGRRRSDDGTVLLHVP
MAB_4701|M.abscessus_ATCC_1997 TIYVGLDAWDARAEALGGASHTLAAAFAARFGERIGRRRAHDGAITLQIP
: : : ** * *.* * :* ..:*:::.: :** **:: : :*
MMAR_2354|M.marinum_M VSERTNDDTRANAVTAITFDLDPDRARTDLQFIRNETKQSLSALKDTPNE
MUL_1530|M.ulcerans_Agy99 VSERTNDDTRANAVTAITFDLDPDRARTDLQFIRNETKQSLSALKDTPNE
MAV_4881|M.avium_104 VNERVAGDTRANAVTNVDVLLGPAAIGTDLRAIRAAVKHALVRHHDLPDE
MSMEG_4729|M.smegmatis_MC2_155 ISDRAPGDTRGNAAYIVNAHLDPTDVTKDLSGARSAIRQALEAYRQEPDE
MAB_4701|M.abscessus_ATCC_1997 ISDRTENDTRAMALSYARVSIDPVSLTTDLREVRAAIKQTLKTLKESPDE
:.:*. .***. * :.* .** * :::* :: *:*
MMAR_2354|M.marinum_M LLKPLPLVPYTPKWLARKMEVLAMGTSDLPVGCSNLGELSQALTRVDGTG
MUL_1530|M.ulcerans_Agy99 LLKPLPLVPYTPKWLARKMEVLAMGTSDLPVGCSNLGELSQALTRVDGTG
MAV_4881|M.avium_104 RWALLPLVPLIPKRVFRRLLGVVTG-GATTVVSSNLGALDPAANRPDGTD
MSMEG_4729|M.smegmatis_MC2_155 TLQLLPLTPFIPQRAVARGSQVVLNLTDLPVSCSNLGDIDPLVSRIDGAA
MAB_4701|M.abscessus_ATCC_1997 SKLLLWLPSFAPERTLKRMVARMPADPDQSVFCSYLGDLHSMIGWPAGTL
* * . *: : .* .* ** : *:
MMAR_2354|M.marinum_M ARYSSARMFNRGATRQRFERESGELYLFSGRLENSVFISVSAYQPGAENS
MUL_1530|M.ulcerans_Agy99 ARYSSARMFNRGATRQRFERESGELYLFSGRLENSVFISVSAYQPGAENS
MAV_4881|M.avium_104 ADYFAMRSVYPGVTDATMQRTNGALALLSGRVHDRVFVSVLGYELGRQNS
MSMEG_4729|M.smegmatis_MC2_155 AEYLILRGVDRRVSREFLENRNGLLTVLAGRVDDKMSISVIGYRPGGPND
MAB_4701|M.abscessus_ATCC_1997 ADFQNARATGQRERRQLLERTGGRMFILSGRLNGKIFISVGAYQPGATNT
* : * ::. .* : :::**:.. : :** .*. * *
MMAR_2354|M.marinum_M NQYLRGLVEQTLADYSLKAELLG---------
MUL_1530|M.ulcerans_Agy99 NQYLRGLVEQTLADYSLKAELLG---------
MAV_4881|M.avium_104 NEALRQALSGALNDFSLTGATGWNPVRAVATR
MSMEG_4729|M.smegmatis_MC2_155 KPQLRELVAKALAEFDLVGTIA----------
MAB_4701|M.abscessus_ATCC_1997 KEALRELAELTLADFSLPGEIH----------
: ** :* ::.* .