For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTTLLRYLGVRLALIIPTAWILMTLVFVLMRGIGDPITAQAGGRLTPAEIAKRKAEAGFDRPIWAQYWE YLTGLLRGDLGQTLTDKRSIADIIVVNGAATLELAVGAIVVALLVGIPLGRIAATHRDRIADVFLRLAAV FFYAAPVFFVAILLKLFFSVKLGWLPASGRSTVGTEIALDAMPHRTHLLLVDTVLYGNGAYFVDALKHLV LPVLALGLLTAGVFLRLVRVNLLQTLRAGYVDAARARGLPERVVVGRHAFRNSLVPVVTVMGMQIAMLLA GAVLTETAFEWNGLGSQLAHYLSARDFIAVQGIVTVIAIIVAVMSFVIDLVVAFIDPRVRF
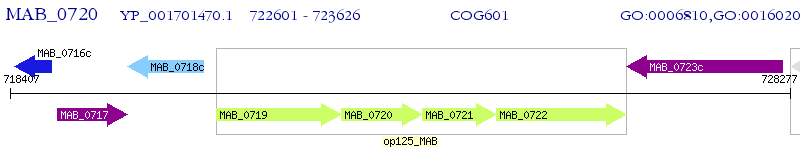
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0720 | - | - | 100% (341) | oligopeptide ABC transporter, permease protein |
| M. abscessus ATCC 19977 | MAB_0427 | - | 9e-42 | 31.59% (345) | peptide ABC transporter transmembrane protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 5e-06 | 24.42% (258) | peptide ABC transporter DppC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3689c | dppB | 3e-37 | 30.38% (339) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2760 | - | 6e-53 | 32.94% (337) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3665c | dppB | 3e-37 | 30.38% (339) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01124 | - | 4e-29 | 26.00% (300) | putative binding-protein dependent transport protein |
| M. marinum M | MMAR_5153 | dppB | 4e-38 | 31.27% (339) | dipeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_0465 | dppB | 4e-41 | 32.45% (339) | ABC transporter, permease protein DppB |
| M. smegmatis MC2 155 | MSMEG_6866 | - | 9e-53 | 34.65% (329) | dipeptide transport system permease protein DppB |
| M. thermoresistible (build 8) | TH_2645 | - | 3e-52 | 33.23% (337) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4239 | dppB | 2e-38 | 31.56% (339) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3774 | - | 3e-53 | 34.40% (343) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2760|M.gilvum_PYR-GCK ------------------MP-------ILRFVARRLLYSAVVMFGVLLVV
Mvan_3774|M.vanbaalenii_PYR-1 -------------MS-TAMP-------VLRFVARRLLFSAVVMIGVLVVV
TH_2645|M.thermoresistible__bu ------------MRF-VTHP-------IARFLGRRLLYSLAVLFGVLVVV
MSMEG_6866|M.smegmatis_MC2_155 MTDLLAGPAPIRAAA-QRHPGGTRALNMGRVVLGKVASALIVLFVVATVA
MAB_0720|M.abscessus_ATCC_1997 -------------MSTT----------LLRYLGVRLALIIPTAWILMTLV
Mb3689c|M.bovis_AF2122/97 ---------------------------MGWYVARRVAVMVPVFLGATLLI
Rv3665c|M.tuberculosis_H37Rv ---------------------------MGWYVARRVAVMVPVFLGATLLI
MAV_0465|M.avium_104 ---------------------------MGWYIARRIAVMVPVFLGATLLI
MMAR_5153|M.marinum_M ---------------------------MVWYVARRILAMVPVFLGATLLI
MUL_4239|M.ulcerans_Agy99 ---------------------------MVWYVARRILAMVPVFLGATLLI
MLBr_01124|M.leprae_Br4923 ---------------------------MMRFLARRLLNYVVLLALASFLT
: : :: :
Mflv_2760|M.gilvum_PYR-GCK FALVHLVPGDPVRIALGTR-YTPEAYEALRAASGLDRPLVSQFFGYIGSA
Mvan_3774|M.vanbaalenii_PYR-1 FALVHLVPGDPVRIALGTR-YTPEAYEALRAASGLDRPIVSQFFGYLGSA
TH_2645|M.thermoresistible__bu FGLMQLVPGDPVRIALGTR-YTPEAYQALRSASGLDRPILEQFVGYLTSA
MSMEG_6866|M.smegmatis_MC2_155 FLLVRMS-GDPLALLLPPD-ATQQQVEQLAAELGLDQPLLVQYLHYLSGM
MAB_0720|M.abscessus_ATCC_1997 FVLMRGI-GDPITAQAGGR-LTPAEIAKRKAEAGFDRPIWAQYWEYLTGL
Mb3689c|M.bovis_AF2122/97 YGMVFLLPGDPVAALAGDRPLTPAVAAQLRSHYHLDDPFLVQYLRYLGGI
Rv3665c|M.tuberculosis_H37Rv YGMVFLLPGDPVAALAGDRPLTPAVAAQLRSHYHLDDPFLVQYLRYLGGI
MAV_0465|M.avium_104 YAMVFLLPGDPVAAIAGDRPLTPAVAAALRARYHLDDPFLVQYLRYLGGV
MMAR_5153|M.marinum_M YGMVFLLPGDPLAAIAGDRPLTPAVAQQLRARYHLDDPFLVQYAHYLGGV
MUL_4239|M.ulcerans_Agy99 YGMVFLLPGDPLAAIAGDRPLTPAVAQQLRARYHLDDPFLVQYAHYLGGV
MLBr_01124|M.leprae_Br4923 YCLTSMAFAPLDSLLQRSPHPPQAMVDAKAHALGLDKPIPIRYANWASHA
: : . . :* *: :: :
Mflv_2760|M.gilvum_PYR-GCK LTGDLGVSFRNGDPVTVTLLERLPATLSLGLAGIVIALAIALPAGIYAAL
Mvan_3774|M.vanbaalenii_PYR-1 LTGDLGVSFRNGDPVTLTLLERLPATLSLGLVGIVIALAIALPAGIYAAL
TH_2645|M.thermoresistible__bu VRGDLGVSFRNGDPVTLVLLERLPATVSLALAGIAIALAIALPAGIWSAL
MSMEG_6866|M.smegmatis_MC2_155 LHGDLGQSIFFDQPVVSLIGERLPATALLAVAALVLSLVISLPAGILASA
MAB_0720|M.abscessus_ATCC_1997 LRGDLGQTLTDKRSIADIIVVNGAATLELAVGAIVVALLVGIPLGRIAAT
Mb3689c|M.bovis_AF2122/97 LHGDLGRAYS-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGL
Rv3665c|M.tuberculosis_H37Rv LHGDLGRAYS-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGL
MAV_0465|M.avium_104 LRGDLGRAYS-GLPVSDVLAHAFPVTLRLSLIALAVEAVLGIGFGVIAGL
MMAR_5153|M.marinum_M LHGDLGRAYS-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGL
MUL_4239|M.ulcerans_Agy99 LHGDLGRAYS-GLPVSAVLAHAFPVTIRLAWIALAVEAVLGIGFGVIAGL
MLBr_01124|M.leprae_Br4923 IRGDFGTTIT-GQPVGSTLGRRVGVSLRLLIIGSLTGTVLGVTAGAWGAI
: **:* : .: : .: * . :.: * ..
Mflv_2760|M.gilvum_PYR-GCK REGRLSDALVRISSQFGVSIPDFWMGILLIA-----LFSTVLGWLPTSGY
Mvan_3774|M.vanbaalenii_PYR-1 REGRLSDALVRISSQFGVSIPDFWMGILLIA-----LFSTVLGWLPTSGY
TH_2645|M.thermoresistible__bu RDGRAGAAAVRVASQVGVSVPDFWMGILLIA-----LFASTLGWLPTSGY
MSMEG_6866|M.smegmatis_MC2_155 HRGRITDTVISGIVLIGQSTPAFWIGILFIL-----LFAVRLRVLPAAGY
MAB_0720|M.abscessus_ATCC_1997 HRDRIADVFLRLAAVFFYAAPVFFVAILLKL-----FFSVKLGWLPASGR
Mb3689c|M.bovis_AF2122/97 RQGGIFDSAVLVTGLVIIAIPIFVLGFLAQF-----LFGVQLEIAPVTVG
Rv3665c|M.tuberculosis_H37Rv RQGGIFDSAVLVTGLVIIAIPIFVLGFLAQF-----LFGVQLEIAPVTVG
MAV_0465|M.avium_104 RQGGLFDSAVLITGLVIIAVPIFVLGFLAQF-----VFGVRLGIAPVTVG
MMAR_5153|M.marinum_M RQGGIFDATVLLTGLIIIAVPIFVLGFLAQY-----LFGVRLGLAPVTVG
MUL_4239|M.ulcerans_Agy99 RQGGIFDATVLLTGLIIIAAPIFVLGFLAQY-----LFGVRLGLAPVTVG
MLBr_01124|M.leprae_Br4923 RQYQLSDRVVTLLALLVLSTPTFVIANLLILSALRVNWAFGIQLFDYTGE
: : . : * * :. * :. : :
Mflv_2760|M.gilvum_PYR-GCK RPL-------------------------FEDPAGWLRHIILPALTVAVVA
Mvan_3774|M.vanbaalenii_PYR-1 RPL-------------------------TEDPAGWLRHVVLPGLTVGVVA
TH_2645|M.thermoresistible__bu RAL-------------------------SDDPVGWLRHLVLPALTVGLVA
MSMEG_6866|M.smegmatis_MC2_155 ---------------------------------GGAAHLVLPAITLAIYS
MAB_0720|M.abscessus_ATCC_1997 STVGTEIALDAMPHRTHLLLVDTVLYGNGAYFVDALKHLVLPVLALGLLT
Mb3689c|M.bovis_AF2122/97 ERA-------------------------------SVGRLLLPGIVLGAMS
Rv3665c|M.tuberculosis_H37Rv ERA-------------------------------SVGRLLLPGIVLGAMS
MAV_0465|M.avium_104 NAA-------------------------------TFTRLLLPGIVLGSVS
MMAR_5153|M.marinum_M ART-------------------------------TLDRLLLPGMVLGAVS
MUL_4239|M.ulcerans_Agy99 ART-------------------------------TLDRLLLPGMVLGAVS
MLBr_01124|M.leprae_Br4923 TSPG-------------------VYGGAWAHVIDRLRHLILPTLTLALVG
:::** :.:.
Mflv_2760|M.gilvum_PYR-GCK AAIMTRYVRAAVLEVASMGYVRTARSKGLTPRVVTFRHTVRNALVPILTI
Mvan_3774|M.vanbaalenii_PYR-1 AAIMTRYVRSAVLEVAAMGYVRTARSKGLSPRIVTFRHTVRNALVPILTI
TH_2645|M.thermoresistible__bu AAIMTRYIRSAVLEVAAAGHVRTARSKGLSPWVLTVRHTVRNALIPVLTI
MSMEG_6866|M.smegmatis_MC2_155 IAVVARVLRTSLAEELSADYIRTATAKGLSRRQVVVGHGLRNASLPVVTV
MAB_0720|M.abscessus_ATCC_1997 AGVFLRLVRVNLLQTLRAGYVDAARARGLPERVVVGRHAFRNSLVPVVTV
Mb3689c|M.bovis_AF2122/97 FAYVVRLTRSAVAANAHADYVRTATAKGLSRPRVVTVHILRNSLIPVVTF
Rv3665c|M.tuberculosis_H37Rv FAYVVRLTRSAVAANAHADYVRTATAKGLSRPRVVTVHILRNSLIPVVTF
MAV_0465|M.avium_104 FAYVVRLTRSTVAVNAHADYVRTATAKGLSRPRVVTVHILRNSLIPVVTF
MMAR_5153|M.marinum_M FAYVVRLTRSAVAANAHADYVRTATAKGLSRPRVVTVHILRNSLIPVVTF
MUL_4239|M.ulcerans_Agy99 FAYVVRLTRSAVAANAHADYVRTATAKGLSRPRVVTVHILRNSLIPVVTF
MLBr_01124|M.leprae_Br4923 AAGYSRYQRNAMLDVLSQDFIRAARAKGLTRRRALLKHGLRTALIPMATL
. * * : ..: :* ::**. * .*.: :*: *.
Mflv_2760|M.gilvum_PYR-GCK TGIQLATLLGGVIVVEVVFAWPGLGRLVFNAVAARDYPLIQGAVLLIAAL
Mvan_3774|M.vanbaalenii_PYR-1 TGIQLATLLGGVIVVEVVFAWPGLGRLVFNAVAARDYPLIQGAVLLIAAL
TH_2645|M.thermoresistible__bu TGIQLATILGGVIVVEVVFAWPGLGRLVYNAVAARDYPVIQGAVLLIAAL
MSMEG_6866|M.smegmatis_MC2_155 VGLEFGSLLGGAILTEQVFSWPGIGRLTVEAISNRDFALVQGAVLFFALV
MAB_0720|M.abscessus_ATCC_1997 MGMQIAMLLAGAVLTETAFEWNGLGSQLAHYLSARDFIAVQGIVTVIAII
Mb3689c|M.bovis_AF2122/97 LGADLGALMGGAIVTEGIFNIHGVGGVLYQAVTRQETPTVVSIVTVLVLI
Rv3665c|M.tuberculosis_H37Rv LGADLGALMGGAIVTEGIFNIHGVGGVLYQAVTRQETPTVVSIVTVLVLI
MAV_0465|M.avium_104 LGADLGALMGGAIVTEGIFNIHGVGGVLYQAVTRQEAPTVVSIVTVLVLI
MMAR_5153|M.marinum_M LGADLGALMGGAVVTEGIFNIHGVGGVLYQAVTRQEAPTVVSIVTVLVMV
MUL_4239|M.ulcerans_Agy99 LGADLGALMGGAVVTEGIFNIHGVGGVLYQAVTRQEAPTVVSIVTVLVMV
MLBr_01124|M.leprae_Br4923 FAYGVAGLVTGALFVEKIFGWHGMGEWMVQGVATQDTNIIAAITIFFGTV
. .. :: *.:..* * *:* . :: :: : . . .: :
Mflv_2760|M.gilvum_PYR-GCK FLLINLLVDVLYAVVDPRIRLG-
Mvan_3774|M.vanbaalenii_PYR-1 FLVINLLVDVLYAVVDPRIRLG-
TH_2645|M.thermoresistible__bu FLLINLLVDLLYAIVDPRIRLA-
MSMEG_6866|M.smegmatis_MC2_155 FVVVNLLVDLSYTLLDPRVRLGR
MAB_0720|M.abscessus_ATCC_1997 VAVMSFVIDLVVAFIDPRVRF--
Mb3689c|M.bovis_AF2122/97 YLITNLLVDLLYAALDPRIRYG-
Rv3665c|M.tuberculosis_H37Rv YLITNLLVDLLYAALDPRIRYG-
MAV_0465|M.avium_104 YLVTNLVVDLLYAVLDPRIRYG-
MMAR_5153|M.marinum_M YLLTNLLVDLLYAALDPRIRYG-
MUL_4239|M.ulcerans_Agy99 YLLTNLLVDLLYAALDPRIRYG-
MLBr_01124|M.leprae_Br4923 ILLAGLLSDVIYAALDPRVRVS-
: .:: *: : :***:*