For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVQAQHPHLPDGIDFTDPELFVHGIPERELAELRHTEPIWWNHTERGVAGFDDDGFWVVSKHKDVKEVSL RCEVFSSEQNTAIPRYLPTTPRERIDATRLIMLNMDPPRHSRLRHIISRGFTPRAISRLRDDLNARAQGI ARAAAQLRHGDFVEQVACELPLQAIAGLMGTPLDEREQLFDWSNRLVGSSDGEDDSAVASAELLMYAMGV AARKTAEPGADICTDLVNADIDGQKLSDDEFGFFVMLLAVAGNETTRNSITHGMHAFTQFPEQWELYKKT RPETAADEIVRWATPVTSFQRTALEDTELGGVRIKKGQRVVMMYRSANFDEEVFENPFTFDIMRDPNPHV GFGGNGEHHCVGANLARMTINLMFNAIADHMPDLASAGEPDRLRSGWLNGVKHWEVDFCPAGYGRAS
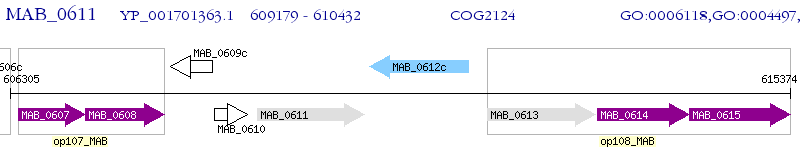
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0611 | - | - | 100% (417) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_0613 | - | e-170 | 69.76% (410) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_1211c | - | e-126 | 51.98% (404) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3575c | cyp125 | 1e-157 | 64.25% (414) | cytochrome P450 125 |
| M. gilvum PYR-GCK | Mflv_3290 | - | 1e-168 | 68.12% (414) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv3545c | cyp125 | 1e-157 | 64.25% (414) | cytochrome P450 125 |
| M. leprae Br4923 | MLBr_02088 | - | 7e-32 | 29.19% (298) | putative cytochrome p450 |
| M. marinum M | MMAR_2783 | cyp125A6 | 1e-166 | 67.90% (405) | cytochrome P450 125A6 Cyp125A6 |
| M. avium 104 | MAV_2811 | - | 1e-166 | 68.15% (405) | putative cytochrome P450 124 |
| M. smegmatis MC2 155 | MSMEG_3524 | - | 1e-163 | 67.33% (404) | linalool 8-monooxygenase |
| M. thermoresistible (build 8) | TH_1956 | - | 1e-157 | 64.44% (405) | PROBABLE CYTOCHROME P450 125 CYP125 |
| M. ulcerans Agy99 | MUL_4106 | cyp125A7 | 1e-161 | 66.18% (414) | cytochrome P450 125A7 Cyp125A7 |
| M. vanbaalenii PYR-1 | Mvan_3012 | - | 1e-165 | 66.83% (416) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2783|M.marinum_M --------MPAAEPTATSVPNLPPGFDFTDPDIYAERLPVAELAEMRRSA
MAV_2811|M.avium_104 --------MATVEPTTKPVPNLPPGFDFTDPDIYAERLPVEELAEMRRVA
MSMEG_3524|M.smegmatis_MC2_155 ------------MVMSDSALHLPAGFDFTDPDIYAERLPVDELAELRRVA
TH_1956|M.thermoresistible__bu ----------------MPTPNIPPGFDFTDPDIYAVRLPVEELAELRRSA
Mflv_3290|M.gilvum_PYR-GCK ----------------MATPTLPPGFDFTDPDLNRERLPVEELAELRRCA
Mvan_3012|M.vanbaalenii_PYR-1 ----------------MATPTLPPGFDFTDPDLNLERLPVEELAELRRCA
Mb3575c|M.bovis_AF2122/97 MSWNHQSVEIAVRRTTVPSPNLPPGFDFTDPAIYAERLPVAEFAELRSAA
Rv3545c|M.tuberculosis_H37Rv MSWNHQSVEIAVRRTTVPSPNLPPGFDFTDPAIYAERLPVAEFAELRSAA
MUL_4106|M.ulcerans_Agy99 ----------------MPCPNLPPGFDFTDPDIYAERLPVEEFAELRSSE
MAB_0611|M.abscessus_ATCC_1997 -------------MVQAQHPHLPDGIDFTDPELFVHGIPERELAELRHTE
MLBr_02088|M.leprae_Br4923 --MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRA
:. . * : : .: :
MMAR_2783|M.marinum_M ---PIWWNEQPTG-CGGFDDGGFWVVTKHKDVKEISLRSDVFSSLQKTAL
MAV_2811|M.avium_104 ---PIWWNEQPIG-AGGFDDGGFWVVTKHKDVKEVSLRSDVFSSLQKTAL
MSMEG_3524|M.smegmatis_MC2_155 ---PIWWNAQPIG-AGGFDDGGFWVVTKHKDVKEISLRSDVFSSLEKTAL
TH_1956|M.thermoresistible__bu ---PIWWNEQPMG-AGGFDDGGFWLVTRHSDVKEVSRRSDVFSSQRKTAL
Mflv_3290|M.gilvum_PYR-GCK ---PIWWNEQPDG-IGGFGDGGFWVVTKHHDVKEISKRSDVFSSEVKTAL
Mvan_3012|M.vanbaalenii_PYR-1 ---PIWWNEQTSGGAGPFGDGGYWVVTKHRDVKEISKHSEVFSSQQKTAL
Mb3575c|M.bovis_AF2122/97 ---PIWWNGQDPGKGGGFHDGGFWAITKLNDVKEISRHSDVFSSYENGVI
Rv3545c|M.tuberculosis_H37Rv ---PIWWNGQDPGKGGGFHDGGFWAITKLNDVKEISRHSDVFSSYENGVI
MUL_4106|M.ulcerans_Agy99 ---PIWWDEQFPGQGGGFHDGGFWAITKLKDVKEVSRRSDVFSSYENGVI
MAB_0611|M.abscessus_ATCC_1997 ---PIWWNHTERG-VAGFDDDGFWVVSKHKDVKEVSLRCEVFSSEQNTAI
MLBr_02088|M.leprae_Br4923 DPFPVYRALIDYG-PMQLPGMPLTVFSSFSDCDEALRHPLSASDRLKATL
*:: * : . .: * .* : *. : .:
MMAR_2783|M.marinum_M PRYKDGTVDEQIERGKFVLLNMDAPQHTRLRKIVSRAFTPRAVERLRDDL
MAV_2811|M.avium_104 PRYKDGTVAEQVERGKFVLLNMDAPQHTRLRKIISRAFTPRAVERLRDDL
MSMEG_3524|M.smegmatis_MC2_155 PRYPEGTVQDQIEQGRFVLLNMDAPHHTHLRKIISRAFTPRAVERLRDDL
TH_1956|M.thermoresistible__bu PRYRDGTVAEQIDRGKYVLLNMDAPHHTRLRKIISRAFTPRAVERLRDEL
Mflv_3290|M.gilvum_PYR-GCK PRYPEGSTGEQIETGSLVLLNMDAPRHTHLRKIISRGFTPRAVERLRDDL
Mvan_3012|M.vanbaalenii_PYR-1 PRYPEGSTTEQVETGSLVLLNMDAPRHTHLRKIISRGFTPRAVERLREDL
Mb3575c|M.bovis_AF2122/97 PRFKNDIAREDIEVQRFVMLNMDAPHHTRLRKIISRGFTPRAVGRLHDEL
Rv3545c|M.tuberculosis_H37Rv PRFKNDIAREDIEVQRFVMLNMDAPHHTRLRKIISRGFTPRAVGRLHDEL
MUL_4106|M.ulcerans_Agy99 PRFKNDIAREDIDVQRFVMLNMDAPHHTRLRKIISRGFTPRAIGRLHDEL
MAB_0611|M.abscessus_ATCC_1997 PRYLPTTPRERIDATRLIMLNMDPPRHSRLRHIISRGFTPRAISRLRDDL
MLBr_02088|M.leprae_Br4923 AQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQALEGDI
.: : :: :*.* *::**:::*:.*:*:.: *. ::
MMAR_2783|M.marinum_M RERARRIVEAAAAEGRGDFVEQVSCELPLQAIAGLMGVPQEDRKKLFHWS
MAV_2811|M.avium_104 RERARRIVEAAAAEGSGDFVEQVSCELPLQAIASLMGVPQEDRKKLFHWS
MSMEG_3524|M.smegmatis_MC2_155 AERARAIVRAAAEEGSGDFVEQVACELPLQAIAGLMGVPQEDRRKLFDWS
TH_1956|M.thermoresistible__bu NERAHRIVMEAAAAGSGDFVEQVACELPLQAIAGLMGVPQEDRKKLFDWS
Mflv_3290|M.gilvum_PYR-GCK NARAQNIAKTAASAGSGDFVEQVSCELPLQAIAGLLGVPVDDRKKLFDWS
Mvan_3012|M.vanbaalenii_PYR-1 AQRAHNIAKSAAAAGAGDFVEQVSCELPLQAIAGLLGVPLEDRKKLFDWS
Mb3575c|M.bovis_AF2122/97 QERAQKIAAEAAAAGSGDFVEQVSCELPLQAIAGLLGVPQEDRGKLFHWS
Rv3545c|M.tuberculosis_H37Rv QERAQKIAAEAAAAGSGDFVEQVSCELPLQAIAGLLGVPQEDRGKLFHWS
MUL_4106|M.ulcerans_Agy99 NDRAQNIAKAAAAAGSGDFVEQVSCELPLQAIAGLLGIPQEDRGKLFDWS
MAB_0611|M.abscessus_ATCC_1997 NARAQGIARAAAQLRHGDFVEQVACELPLQAIAGLMGTPLDEREQLFDWS
MLBr_02088|M.leprae_Br4923 AALVDSLLDKGAAAGQFDVIADLAFPLAVAVICRLLGVPYEDAPEFGRVS
. : .* *.: ::: *.: .*. *:* * :: :: *
MMAR_2783|M.marinum_M NEMVG--------DQDPEFATNDALTASVELIMYGMQMAADRAKNPGQDL
MAV_2811|M.avium_104 NEMVG--------DQDPEFASNDAITASVELIMYGMQMAADRAKNPGEDL
MSMEG_3524|M.smegmatis_MC2_155 NQMVG--------NQDPEFVANDGASAAVELITYGMQLAAQRAAAPGDDL
TH_1956|M.thermoresistible__bu NQMVG--------DQDPEFADNDAIGASIELIGYGMQLAADRQRNPRDDL
Mflv_3290|M.gilvum_PYR-GCK NQMVS--------DDDPEYAHYDNRNAATELIMYAMQLAAVRAEQPGEDI
Mvan_3012|M.vanbaalenii_PYR-1 NQMVS--------DDDPEFAHYDNRNAATELIMYAMQLAALRAEQPGEDI
Mb3575c|M.bovis_AF2122/97 NEMTG--------NEDPEYAHIDPKASSAELIGYAMKMAEEKAKNPADDI
Rv3545c|M.tuberculosis_H37Rv NEMTG--------NEDPEYAHIDPKASSAELIGYAMKMAEEKAKNPADDI
MUL_4106|M.ulcerans_Agy99 NEMTG--------TEDPEFAHIDAKASSVELIGYAMKMAEEKAKNPGDDI
MAB_0611|M.abscessus_ATCC_1997 NRLVG--------SSD---GEDDSAVASAELLMYAMGVAARKTAEPGADI
MLBr_02088|M.leprae_Br4923 ALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRGTPGEDL
:. : : :. * * :. : * *:
MMAR_2783|M.marinum_M VTKLVEADIDGHKLSDDEFGFFVILLAVAGNETTRNSITQGMMAFTDFPD
MAV_2811|M.avium_104 VTKLVQADIDGHKLSDDEFGFFVILLAVAGNETTRNSITQGMMAFTDFPD
MSMEG_3524|M.smegmatis_MC2_155 VTKLVQADVEGHKLSDDEFGFFVVLLAVAGNETTRNSITQGMMAFTDHRD
TH_1956|M.thermoresistible__bu VTTLVQADVDGHKLSDDEFGFFVILLAVAGNETTRNSITQGMMAFTEHPD
Mflv_3290|M.gilvum_PYR-GCK VTKLIEADVDGHKLSDDEFGFFMVLLAVAGNETTRNSITHGMIAFTEHPD
Mvan_3012|M.vanbaalenii_PYR-1 VTKLIEADVDGHKLTDDEFGFFMVLLAVAGNETTRNSITHGMIAFTEHPD
Mb3575c|M.bovis_AF2122/97 VTQLIQADIDGEKLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFAEHPD
Rv3545c|M.tuberculosis_H37Rv VTQLIQADIDGEKLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFAEHPD
MUL_4106|M.ulcerans_Agy99 VTQLIQADIDGEKLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFADNPE
MAB_0611|M.abscessus_ATCC_1997 CTDLVNADIDGQKLSDDEFGFFVMLLAVAGNETTRNSITHGMHAFTQFPE
MLBr_02088|M.leprae_Br4923 ISRLIELDESGDQLTEEEIIATCGLLLVAGHETTVNLIANAVLAMLRNPS
: *:: * .*.:*:::*: :* ***:*** * *::.: *: .
MMAR_2783|M.marinum_M QWELYKR--ERPVTTADEIVRWATPVTSFQRTALQDYELSGVRIKKGQRV
MAV_2811|M.avium_104 QWELFKR--ERPATAADEIVRWATPVTSFQRTALQDYELSGVKIKKGQRV
MSMEG_3524|M.smegmatis_MC2_155 QWELFKR--ERPVTTADEIVRWATPVTSFQRTALADTEVSGVRIKKGQRV
TH_1956|M.thermoresistible__bu QWELFKE--QRPPTTADEIVRWATPVTSFQRTALADVELSGVQIKAGQRV
Mflv_3290|M.gilvum_PYR-GCK QWELYKR--ERPITAVDEIVRWATPVTSFQRTALTDYELSGVQITKGQRV
Mvan_3012|M.vanbaalenii_PYR-1 QWELFKR--ERPATAVDEIVRWATPVTSFQRTALRDYELSGVQIKKGQRV
Mb3575c|M.bovis_AF2122/97 QWELYKK--VRPETAADEIVRWATPVTAFQRTALRDYELSGVQIKKGQRV
Rv3545c|M.tuberculosis_H37Rv QWELYKK--VRPETAADEIVRWATPVTAFQRTALRDYELSGVQIKKGQRV
MUL_4106|M.ulcerans_Agy99 QWELYKR--ERPGTAADEIVRWATPVTSFQRTALEDYELSGVQIKKGQRV
MAB_0611|M.abscessus_ATCC_1997 QWELYKK--TRPETAADEIVRWATPVTSFQRTALEDTELGGVRIKKGQRV
MLBr_02088|M.leprae_Br4923 QWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKDMTIGQTTLTEGDTM
**: . *. ..:* :*: ..: : *.* * :. . :. *: :
MMAR_2783|M.marinum_M VMFYRSANFDEDVFDDPYTFNILRDPNPHVGFGGTGAHYCIGANLARMTI
MAV_2811|M.avium_104 VMFYRSANFDEDVFDDPFTFNILRDPNPHVGFGGTGAHYCIGANLARMTI
MSMEG_3524|M.smegmatis_MC2_155 VMFYRSANFDEDVFTDPYRFDILRDPNPHVGFGGTGAHYCIGANLARMTI
TH_1956|M.thermoresistible__bu VMCYRSANFDDTVFDDPYTFNILRDPNPHLGFGGTGAHYCIGANLARLTI
Mflv_3290|M.gilvum_PYR-GCK VMSYRSANFDEEVFDDPFTFDILRDPNPHVGFGGTGAHYCIGANLARMTI
Mvan_3012|M.vanbaalenii_PYR-1 VMSYRSANFDEEVFDDPFTFDIMRDPNPHVGFGGTGAHYCIGANLARMTI
Mb3575c|M.bovis_AF2122/97 VMFYRSANFDEEVFQDPFTFNILRNPNPHVGFGGTGAHYCIGANLARMTI
Rv3545c|M.tuberculosis_H37Rv VMFYRSANFDEEVFQDPFTFNILRNPNPHVGFGGTGAHYCIGANLARMTI
MUL_4106|M.ulcerans_Agy99 LMFYRSANFDEEVFEDPFSFNILRNPNPHVGFGGTGAHYCIGANLARMTI
MAB_0611|M.abscessus_ATCC_1997 VMMYRSANFDEEVFENPFTFDIMRDPNPHVGFGGNGEHHCVGANLARMTI
MLBr_02088|M.leprae_Br4923 VLLLAAANRDPAVYSRPDEFDPDRPSSRHLAFA-VGSHFCLGAALARLEA
:: :** * *: * *: * .. *:.*. * *.*:** ***:
MMAR_2783|M.marinum_M DLMFNAIADVMPDLESISQPERLRSGWLNGIKHWQVDYHSDSSGKCPVAH
MAV_2811|M.avium_104 DLMFNAIADAMPDLESIGKPERLRSGWLNGIKHWQVDYHTNGSSKCPVAH
MSMEG_3524|M.smegmatis_MC2_155 DLIFNAIADEMPDLTPISEPVRLRSGWLNGIKHWQVDYRGDAAKHRAAQA
TH_1956|M.thermoresistible__bu DLIFAAIAEHLPDLRPLGPPERLRSGWLNGIKHWPVDYGAPRVPTGAAGP
Mflv_3290|M.gilvum_PYR-GCK DLMFNAIADHMPDLSAIGSPDRLRSGWLNGIKHWQVDYSG---RGCPVAH
Mvan_3012|M.vanbaalenii_PYR-1 DLMFNAIADHLPDLSSAGTPDRLRSGWLNGIKHWQVDYTGP--SGCPVAH
Mb3575c|M.bovis_AF2122/97 NLIFNAVADHMPDLKPISAPERLRSGWLNGIKHWQVDYTG----RCPVAH
Rv3545c|M.tuberculosis_H37Rv NLIFNAVADHMPDLKPISAPERLRSGWLNGIKHWQVDYTG----RCPVAH
MUL_4106|M.ulcerans_Agy99 NLIFNAVADHMPDLKPIAAPERLRSGWLNGIKHWQVDYTG----KCPVSH
MAB_0611|M.abscessus_ATCC_1997 NLMFNAIADHMPDLASAGEPDRLRSGWLNGVKHWEVDFCPAGYGRAS---
MLBr_02088|M.leprae_Br4923 TVTLSAISARFPQVQLAGELVYKPNVAMRGMSALPVQV------------
: : *:: :*:: . . :.*:. *:
MMAR_2783|M.marinum_M ------
MAV_2811|M.avium_104 ------
MSMEG_3524|M.smegmatis_MC2_155 SSQADR
TH_1956|M.thermoresistible__bu QRCG--
Mflv_3290|M.gilvum_PYR-GCK ------
Mvan_3012|M.vanbaalenii_PYR-1 ------
Mb3575c|M.bovis_AF2122/97 ------
Rv3545c|M.tuberculosis_H37Rv ------
MUL_4106|M.ulcerans_Agy99 ------
MAB_0611|M.abscessus_ATCC_1997 ------
MLBr_02088|M.leprae_Br4923 ------