For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PTRPSQNDPERVVAWNRIRQTVGARIRQIRTSQDMTQEQLALRSGVTRNVLIDVEHGRRGLLYERLFDIA AALEVTVGALLHED*
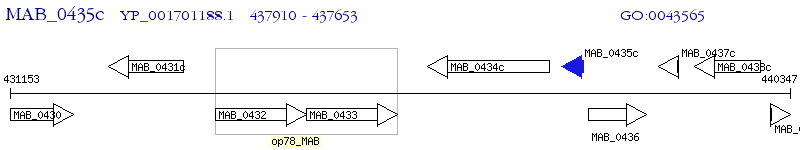
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0435c | - | - | 100% (85) | putative transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_4871 | - | 6e-18 | 87.23% (47) | hypothetical protein MAB_4871 |
| M. abscessus ATCC 19977 | MAB_0769c | - | 2e-06 | 37.93% (58) | putative transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0885 | - | 9e-31 | 75.61% (82) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2152 | - | 3e-05 | 34.92% (63) | transcriptional regulator |
| M. avium 104 | MAV_1459 | - | 5e-15 | 80.85% (47) | transcriptional regulator, XRE family protein |
| M. smegmatis MC2 155 | MSMEG_1151 | - | 3e-05 | 31.67% (60) | DNA-binding protein |
| M. thermoresistible (build 8) | TH_0145 | - | 3e-05 | 33.85% (65) | regulatory protein |
| M. ulcerans Agy99 | MUL_1737 | - | 2e-05 | 34.92% (63) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3268 | - | 5e-27 | 71.95% (82) | XRE family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0885|M.gilvum_PYR-GCK -------------------MPTRPSQNNPERVAQWEHWRRLVGAKIREL-
Mvan_3268|M.vanbaalenii_PYR-1 -------------------MPTRPSQNDPERVSAWAAHRRTVGDNIRRL-
MAB_0435c|M.abscessus_ATCC_199 -------------------MPTRPSQNDPERVVAWNRIRQTVGARIRQI-
MAV_1459|M.avium_104 --------------------------------------------------
MMAR_2152|M.marinum_M -------------------MTALLRAVRRQRGLTLEQLAQRAGLTKSYLS
MUL_1737|M.ulcerans_Agy99 -------------------MTALLRAVRRQRGLTLEQLAQRAGLTKSYLS
TH_0145|M.thermoresistible__bu ------MEANQREGRIEDRIRQRLRELRKQRGLTLEDVATRAQIDVSTLS
MSMEG_1151|M.smegmatis_MC2_155 MGKAKPNDNKALVDAMLGDLGSRIRMLRKERQLTTERLAETAGVSAGLIS
Mflv_0885|M.gilvum_PYR-GCK RIARDMTQEALALRSGVTRNVLIDV------EHGRRGLLYERLFD-----
Mvan_3268|M.vanbaalenii_PYR-1 RISCGLTQEALALQSGVTRNVLLAV------EHGRRGLLYERLFD-----
MAB_0435c|M.abscessus_ATCC_199 RTSQDMTQEQLALRSGVTRNVLIDV------EHGRRGLLYERLFD-----
MAV_1459|M.avium_104 -----MTQEVLALHSGVTRNVLIDV------EFGRRGLLYERLFD-----
MMAR_2152|M.marinum_M KIERGQSTPSIAVALKVARALDVDVGRLFSDESAHETITVDRAHDRLGPQ
MUL_1737|M.ulcerans_Agy99 KIERGQSTPSIAVALKVARALDVDVGRLFSDESAHETITADRAHDRLGPQ
TH_0145|M.thermoresistible__bu RLESGKRRLAIDHLPRLAAALSVSTDDLLRAPDTEDPRVRGSAHTSHG--
MSMEG_1151|M.smegmatis_MC2_155 QIERGNGNPSFATLVQLAHGLQMPIGQLLEAPESKSVVVRKNERRRLDGH
: :: : : .
Mflv_0885|M.gilvum_PYR-GCK ------------IADALDVNVAELLP------------------------
Mvan_3268|M.vanbaalenii_PYR-1 ------------IADALGVPASRLLDSHNE----SGEGLRTADSGDSAQP
MAB_0435c|M.abscessus_ATCC_199 ------------IAAALEVTVGALLHED----------------------
MAV_1459|M.avium_104 ------------LAEALDVPVADLLAVPEP----PPKRKRRSKRSTQ---
MMAR_2152|M.marinum_M -----SGRYHVLASTQLGKTMSPFVVRPTAGQDCASDGPHPEHTGQEFVF
MUL_1737|M.ulcerans_Agy99 -----SGRYHVLASTQLGKTMSPFVVRPTAGQDCASDGPHPEHTGQEFVF
TH_0145|M.thermoresistible__bu ------VTYWPLTRQGPAGGLHAFKIRISARRRTPPAG-LPVHEGHDWVY
MSMEG_1151|M.smegmatis_MC2_155 GLANDDGGSYELLTPDLNGALEATWVVTPPG--YDTSATPYHHHGEEFGI
Mflv_0885|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3268|M.vanbaalenii_PYR-1 L-------------------------------------------------
MAB_0435c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1459|M.avium_104 --------------------------------------------------
MMAR_2152|M.marinum_M VHTGSVELDYGDQTITLGTGDSAYFDASVGHKLRAVGTERAEVVVVAGAE
MUL_1737|M.ulcerans_Agy99 VHTGSVELDYGDQTITLGTGDSAYFDASVGHKLRAVGTERAEVVVVAGAE
TH_0145|M.thermoresistible__bu VLSGHMRLILDSRDFVIKPGEAVEFSTWTPHWFGAVDGPVEAIMILGPHG
MSMEG_1151|M.smegmatis_MC2_155 VLSGTKDVYLDGIRHRLHAGDSIRYASNIPHWYVNPSETEECTAIWVSTP
Mflv_0885|M.gilvum_PYR-GCK -------
Mvan_3268|M.vanbaalenii_PYR-1 -------
MAB_0435c|M.abscessus_ATCC_199 -------
MAV_1459|M.avium_104 -------
MMAR_2152|M.marinum_M LGGR---
MUL_1737|M.ulcerans_Agy99 LGGR---
TH_0145|M.thermoresistible__bu ERLHLHE
MSMEG_1151|M.smegmatis_MC2_155 PTW----