For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DNDQSAPAEVGARLRRLRTEAGLSLAELATRSGVGKGSISELENGRRTARLDTLFALTKALGAPLSAAVG VSPPADGPSVHGQAVHAVSLSGWQTDDGYVEVYRITVETTTQHSPAHATGVRETITVVAGTLEAGPLGDP QIAAAGETMCFPGNVPHVYRAVNGPAEAVLTMHYPPGITTPVAQ*
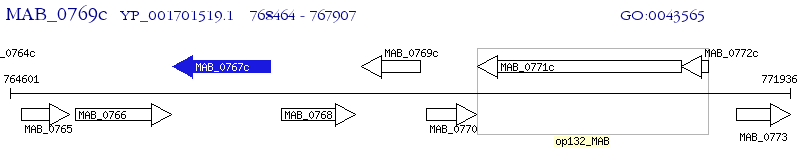
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0769c | - | - | 100% (185) | putative transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_4363c | - | 1e-14 | 32.00% (175) | putative DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_3176c | - | 8e-08 | 27.27% (176) | hypothetical protein MAB_3176c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1295 | - | 2e-07 | 35.14% (74) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1378 | - | 6e-05 | 32.23% (121) | transcriptional regulatory protein |
| M. avium 104 | MAV_2214 | - | 2e-11 | 30.18% (169) | regulatory protein |
| M. smegmatis MC2 155 | MSMEG_4304 | - | 4e-12 | 31.61% (174) | regulatory protein |
| M. thermoresistible (build 8) | TH_0145 | - | 5e-11 | 27.11% (166) | regulatory protein |
| M. ulcerans Agy99 | MUL_2497 | - | 4e-05 | 32.23% (121) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3617 | - | 1e-12 | 29.05% (179) | XRE family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2214|M.avium_104 -------------------------MQAN---TDIDVQVRRRLRELRVQR
TH_0145|M.thermoresistible__bu -------------------------MEANQREGRIEDRIRQRLRELRKQR
Mvan_3617|M.vanbaalenii_PYR-1 -------------------------MKANEEDDGVDARVRRRLRDLRLQR
MSMEG_4304|M.smegmatis_MC2_155 -------------------------MDAPGAEHDVDALVRQRLRELRTER
MMAR_1378|M.marinum_M ---------------------------------MPKTFAGARLRRLREDH
MUL_2497|M.ulcerans_Agy99 -----------------------MKGGARRVSHVPKTFAGARLRRLREDH
MAB_0769c|M.abscessus_ATCC_199 ----------------------------MDNDQSAPAEVGARLRRLRTEA
Mflv_1295|M.gilvum_PYR-GCK MANQNDRDDHTPLLRNTSGTARDRDPGAPVEELEIESAIGRNVRLLRQQQ
.:* ** :
MAV_2214|M.avium_104 GLTLQEVGARAGIDVSTLSRLESGKRRLALDHLPRLARALSVSTDELLQ-
TH_0145|M.thermoresistible__bu GLTLEDVATRAQIDVSTLSRLESGKRRLAIDHLPRLAAALSVSTDDLLR-
Mvan_3617|M.vanbaalenii_PYR-1 GMTLEDVATRSNIDISTLSRLESGKRRLALDHLPRLASALSVSTDELLR-
MSMEG_4304|M.smegmatis_MC2_155 GLTLEDVASRAQIDVSTLSRLESGKRRLALDHLPRLATALSVTTDDLLR-
MMAR_1378|M.marinum_M GLTQVALARALGLSTSYVNQLENDQRPITVSVLLALAERFDLPTHYFAPD
MUL_2497|M.ulcerans_Agy99 GLTQVALARALGLSTSYVNQLENDQRPITVSVLLALAERFDLPTHYFAPD
MAB_0769c|M.abscessus_ATCC_199 GLSLAELATRSGVGKGSISELENGRRTARLDTLFALTKALGAPLSAAVG-
Mflv_1295|M.gilvum_PYR-GCK GLTVAETAARVGISKAMMSKIENAQTSCSLSTLALLAKGFDVPVTSLFRG
*:: . :. . :..:*. : :. * *: :. .
MAV_2214|M.avium_104 -----PAQAPDPRVRG---SAHTHHGVTYWPLTRQ----------GPAGG
TH_0145|M.thermoresistible__bu -----APDTEDPRVRG---SAHTSHGVTYWPLTRQ----------GPAGG
Mvan_3617|M.vanbaalenii_PYR-1 -----APEVEDPRVRG---NSHTHNLITFWPLTGQ----------GPAGG
MSMEG_4304|M.smegmatis_MC2_155 -----APEAEDPRVRG---TATTRDNITYWPLTRH----------GPANG
MMAR_1378|M.marinum_M SDARLVSDLREVFAEGPATPAQIEELVARMPAVGQTLVNLHRRLYDATAE
MUL_2497|M.ulcerans_Agy99 SDARLVSDLREVFAEGPATPAQIEELVARMPAVGQTLVNLHRRLYDATAE
MAB_0769c|M.abscessus_ATCC_199 ----VSPPADGPSVHG---QAVHAVSLSGWQTDDG--------------Y
Mflv_1295|M.gilvum_PYR-GCK ADVERPAAFVKAGTGARIVREGTREGHEYQLLGSLR---------GEHKR
. . .
MAV_2214|M.avium_104 LHAFKIRVSAQRRTPP-AELPVHEGQDWMY--------------------
TH_0145|M.thermoresistible__bu LHAFKIRISARRRTPP-AGLPVHEGHDWVY--------------------
Mvan_3617|M.vanbaalenii_PYR-1 LHAYKIRISARRRTPP-AELPVHEGHDWMY--------------------
MSMEG_4304|M.smegmatis_MC2_155 LHAYKIRISEQRNTPP-AELPVHEGRDWMY--------------------
MMAR_1378|M.marinum_M LEALHSRATADVSAVSGQPMPFEEVRDFFYDRKNYIGELDIAAEEMFGRH
MUL_2497|M.ulcerans_Agy99 LEALHSRATADVSAVSGQPMPFEEVRDFFYDRKNYIGELDIAAEQMFGRH
MAB_0769c|M.abscessus_ATCC_199 VEVYRITVETTTQHSPAHATGVRETITVVAG-------------------
Mflv_1295|M.gilvum_PYR-GCK LECLEVTLSEKSQTYP---LFQHPGTEFIY--------------------
:. . . . .
MAV_2214|M.avium_104 -----VLAGRMRLILGD---RDFTIDPGEAVEFST-----------WTPH
TH_0145|M.thermoresistible__bu -----VLSGHMRLILDS---RDFVIKPGEAVEFST-----------WTPH
Mvan_3617|M.vanbaalenii_PYR-1 -----VLSGRLRLMLGD---RDFTIKPGEAVEFST-----------WTPH
MSMEG_4304|M.smegmatis_MC2_155 -----VLSGKLRLILGE---QDFTVGPGEAVEFST-----------WTPH
MMAR_1378|M.marinum_M GLRLGGLDAQLARLLGDNLGTTVVLDDGQSLNPNSKRMFQPESKTLYLAR
MUL_2497|M.ulcerans_Agy99 GLHLGGLDAQLARLLGDNLGITVVLDDGQSLNPNSKRMFQPESKTLYLAR
MAB_0769c|M.abscessus_ATCC_199 --------TLEAGPLGD----PQIAAAGETMCFPG-----------NVPH
Mflv_1295|M.gilvum_PYR-GCK -----MLEGVMDYSHSR---SVYRLQPGDSLQIDG-------------EG
. *:::
MAV_2214|M.avium_104 WFG----------VVDGPVEAIVIFGPHGERLHLHS--------------
TH_0145|M.thermoresistible__bu WFG----------AVDGPVEAIMILGPHGERLHLHE--------------
Mvan_3617|M.vanbaalenii_PYR-1 WFG----------VVDGPVEAIMLLGLHGERVHLHSDQR-----------
MSMEG_4304|M.smegmatis_MC2_155 WFG----------VVDGPVEAVTIFGLHGEQVHLHS--------------
MMAR_1378|M.marinum_M WLNRGQRAFQLATQIALLTQADLITGIIAGDDQLSDEARGVARIGLANYF
MUL_2497|M.ulcerans_Agy99 WLNRGQRAFQLATQIALLTQADLITGIIAGDDQLSDEARGVARIGLANYF
MAB_0769c|M.abscessus_ATCC_199 VYR----------AVNGPAEAVLTMHYPPGITTPVAQ-------------
Mflv_1295|M.gilvum_PYR-GCK AHG-------PVDLVEVPIRFLSVIAFPDSQV------------------
: .
MAV_2214|M.avium_104 --------------------------------------------------
TH_0145|M.thermoresistible__bu --------------------------------------------------
Mvan_3617|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4304|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1378|M.marinum_M AGALLLPYRPFLDAAASTRYDIDQLARRFEVGFETICHRLSTLQRPNARG
MUL_2497|M.ulcerans_Agy99 AGALLLPYRPFLDAAASTRYDIDQLARRFEVGFETICHRLSTLQRPNARG
MAB_0769c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1295|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2214|M.avium_104 --------------------------------------------------
TH_0145|M.thermoresistible__bu --------------------------------------------------
Mvan_3617|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4304|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1378|M.marinum_M VPFIFVRTDSAGNISKRQSATAFHFSRVGGNCPLWVIHQAFARPGRFLTQ
MUL_2497|M.ulcerans_Agy99 VPFIFVRTDSAGNISKRQSATAFHFSRVGGNCPLWVIHQAFARPGQFLTQ
MAB_0769c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1295|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2214|M.avium_104 --------------------------------------------------
TH_0145|M.thermoresistible__bu --------------------------------------------------
Mvan_3617|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4304|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1378|M.marinum_M VAQMPDERTYFWIARTTTAEPSRYLGPGKSFAIGLGCDLAHADKLIYSVG
MUL_2497|M.ulcerans_Agy99 VAQMPDERTYFWIARTTTAEPSRYLGPGKSFAIGLGCDLAHADKLIYSVG
MAB_0769c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1295|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2214|M.avium_104 --------------------------------------------------
TH_0145|M.thermoresistible__bu --------------------------------------------------
Mvan_3617|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4304|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1378|M.marinum_M VDLTDTEAIAAIGAGCKICDRPSCPQRAFPYLGRPVRVDPHTSTDLPYPP
MUL_2497|M.ulcerans_Agy99 VDLTDTEAIAAIGAGCKICDRPSCPQRAFPYLGRPVRVDPHTSTDLPYPP
MAB_0769c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1295|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2214|M.avium_104 ------
TH_0145|M.thermoresistible__bu ------
Mvan_3617|M.vanbaalenii_PYR-1 ------
MSMEG_4304|M.smegmatis_MC2_155 ------
MMAR_1378|M.marinum_M AISTER
MUL_2497|M.ulcerans_Agy99 AISTER
MAB_0769c|M.abscessus_ATCC_199 ------
Mflv_1295|M.gilvum_PYR-GCK ------