For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPTTGSAAKPRAKSSVKAPAPVRAWKPETHTGLVRRARRMNRTLAQAFPHVYCELDFTNPLELAVATILS AQCTDVRVNMVTPALFAKYRTAEDYAGANRAELEEMIRTTGFYRNKANSIMGLGTQLVERFGGEIPPRLK DLVTLPGIGRKTANVILGNAFDIPGITVDTHFGRLVRRWRWTEEEDPVKVEHLVGELIERKEWTLLSHRV IFHGRRVCHARKPACGVCVLAKDCPSYGLGPTDPEVAAPLVKGPETEHLLALAGL
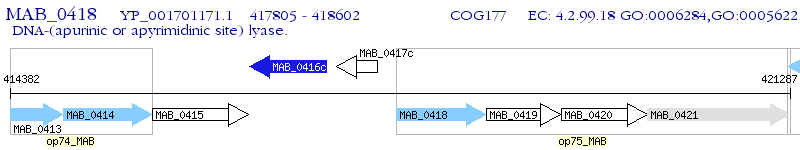
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0418 | - | - | 100% (265) | endonuclease III protein |
| M. abscessus ATCC 19977 | MAB_0558c | - | 1e-05 | 23.96% (192) | adenine glycosylase MutY |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3698c | nth | 1e-116 | 82.16% (241) | endonuclease III |
| M. gilvum PYR-GCK | Mflv_1373 | - | 1e-121 | 79.55% (264) | endonuclease III |
| M. tuberculosis H37Rv | Rv3674c | nth | 1e-116 | 82.16% (241) | endonuclease III |
| M. leprae Br4923 | MLBr_02301 | nth | 1e-113 | 79.84% (243) | putative endonuclease III |
| M. marinum M | MMAR_5162 | nth | 1e-120 | 79.46% (258) | endonuclease III Nth |
| M. avium 104 | MAV_0455 | nth | 1e-103 | 75.43% (232) | endonuclease III |
| M. smegmatis MC2 155 | MSMEG_6187 | nth | 1e-120 | 80.38% (260) | endonuclease III |
| M. thermoresistible (build 8) | TH_2130 | nth | 1e-102 | 81.40% (215) | PROBABLE ENDONUCLEASE III NTH (DNA-(APURINIC OR APYRIMIDINIC |
| M. ulcerans Agy99 | MUL_4248 | nth | 1e-116 | 84.55% (233) | endonuclease III Nth |
| M. vanbaalenii PYR-1 | Mvan_5433 | - | 1e-122 | 85.19% (243) | endonuclease III |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5162|M.marinum_M -----MSGGSKPTANASTPKLPKRWSEETRTALVRRARRMNRKLAQAFPH
MUL_4248|M.ulcerans_Agy99 --------------------------------MVRRARRMNRKLAQAFPH
Mb3698c|M.bovis_AF2122/97 --------------------MPGRWSAETRLALVRRARRMNRALAQAFPH
Rv3674c|M.tuberculosis_H37Rv --------------------MPGRWSAETRLALVRRARRMNRALAQAFPH
MLBr_02301|M.leprae_Br4923 ------------MPLTPGVDVARRWSGETRLGLVRRARRMNRALAQAFPH
MAV_0455|M.avium_104 ---------------------------------MRRARRMNRILAQAFPE
Mflv_1373|M.gilvum_PYR-GCK -----MTVSEPSTGAKSARRSARKWDRETHLGLVRRARRMNRTLTQAFPH
Mvan_5433|M.vanbaalenii_PYR-1 -----MTVK-PARGA-SPRKSGRKWDEETHLGLVRRARRMNRTLAQAFPH
MSMEG_6187|M.smegmatis_MC2_155 -----MSAG-AARAAKPKKSDAKKWDSETHLGLVRRARRMNRTLAKAFPH
TH_2130|M.thermoresistible__bu --------------------------------------------------
MAB_0418|M.abscessus_ATCC_1997 MPTTGSAAKPRAKSSVKAPAPVRAWKPETHTGLVRRARRMNRTLAQAFPH
MMAR_5162|M.marinum_M VYCELDFTSPLELAVATILSAQSTDKRVNLTTPDLFVKYQTALDYAQADR
MUL_4248|M.ulcerans_Agy99 VYCELDFTSPLELAVATILSAQSTDKRVNLTTPDLFAKYQTALDYAQADR
Mb3698c|M.bovis_AF2122/97 VYCELDFTTPLELAVATILSAQSTDKRVNLTTPALFARYRTARDYAQADR
Rv3674c|M.tuberculosis_H37Rv VYCELDFTTPLELAVATILSAQSTDKRVNLTTPALFARYRTARDYAQADR
MLBr_02301|M.leprae_Br4923 VYCELDFTSPLELTVATILSAQSTDKRVNLTTPAVFARYRSALDYMQADR
MAV_0455|M.avium_104 AHCELDFTTPLELTVATILSAQSTDKRVNLTTRALFKRYTCALDYAQADR
Mflv_1373|M.gilvum_PYR-GCK VYCELDFTNPLELTVATILSAQSTDKRVNLTTPALFAKYRTARDYATADR
Mvan_5433|M.vanbaalenii_PYR-1 VYCELDFTDPLELTVATILSAQSTDKRVNLTTPALFKKYRTARDYATADR
MSMEG_6187|M.smegmatis_MC2_155 VYCELDFTNPLELTVATILSAQSTDKRVNLTTPALFKKYRTALDYAQADR
TH_2130|M.thermoresistible__bu VYCELDFTNPLELTVATILSAQSTDKRVNLTTPALFRKYRTALDYAQADR
MAB_0418|M.abscessus_ATCC_1997 VYCELDFTNPLELAVATILSAQCTDVRVNMVTPALFAKYRTAEDYAGANR
.:****** ****:********.** ***:.* :* :* * ** *:*
MMAR_5162|M.marinum_M AELENLIRPTGFYRNKANSLIGLGQALVERFDGQVPATMEELVTLPGVGR
MUL_4248|M.ulcerans_Agy99 AELENLIRPTGFYRNKANSLIGLGQALVERFDGQVPATMEELVTLPGVGR
Mb3698c|M.bovis_AF2122/97 TELESLIRPTGFYRNKAASLIGLGQALVERFGGEVPATMDKLVTLPGVGR
Rv3674c|M.tuberculosis_H37Rv TELESLIRPTGFYRNKAASLIGLGQALVERFGGEVPATMDKLVTLPGVGR
MLBr_02301|M.leprae_Br4923 AELENFIRPTGFFRNKAASLIRLGQALVERFDGEVPSTMVDLFTLPGVGR
MAV_0455|M.avium_104 DELENLIRPTGFFRNKASALIRLGQALVERFDGEVPATMAELVTLPGVGR
Mflv_1373|M.gilvum_PYR-GCK TELEELIRPTGFYRNKATSLIGLGQALEERFDGEVPRTLDELVTLPGIGR
Mvan_5433|M.vanbaalenii_PYR-1 TELEELIRPTGFYRNKANSLIGLGQALEERFDGQVPRTLDELVTLPGVGR
MSMEG_6187|M.smegmatis_MC2_155 TELEELIRPTGFYRNKANSLIKLGQELVERFDGEVPKTLDELVTLPGVGR
TH_2130|M.thermoresistible__bu AELEELIRPTGFFRTKANSLIRLGQELVERFDGEVPDNLEDLVTLPGVGR
MAB_0418|M.abscessus_ATCC_1997 AELEEMIRTTGFYRNKANSIMGLGTQLVERFGGEIPPRLKDLVTLPGIGR
***.:**.***:*.** ::: ** * ***.*::* : .*.****:**
MMAR_5162|M.marinum_M KTANVILGNAFDVPGITVDTHFGRLARRWRWTAEEDPVKVEHAVGELIER
MUL_4248|M.ulcerans_Agy99 KTANVILGNAFDVPGITVDTHFGRLARRWRWTAEEDPVKVEHAVGELIER
Mb3698c|M.bovis_AF2122/97 KTANVILGNAFGIPGITVDTHFGRLVRRWRWTTAEDPVKVEQAVGELIER
Rv3674c|M.tuberculosis_H37Rv KTANVILGNAFGIPGITVDTHFGRLVRRWRWTTAEDPVKVEQAVGELIER
MLBr_02301|M.leprae_Br4923 KTANVILGNAFGIPGITVDTHFGRLVRRWRWTAEEDPVKVEHAVGELIER
MAV_0455|M.avium_104 KTANVILGNAFGVPGITVDTHFARLVHRWRWTAEKDPVKIEHAVGELIER
Mflv_1373|M.gilvum_PYR-GCK KTANVVLGNAFDIPGITVDTHFGRLVRRWRWTAEEDPVKVEHIVGDLIER
Mvan_5433|M.vanbaalenii_PYR-1 KTANVILGNAFDIPGITVDTHFGRLVRRWRWTAEEDPVKVEHIVGELIER
MSMEG_6187|M.smegmatis_MC2_155 KTANVILGNAFDIPGITVDTHFGRLVRRWRWTDHEDPVKVEFAVAELIER
TH_2130|M.thermoresistible__bu KTANVILGNAFDVPGITVDTHFGRLVRRWRWTDERDPVKVEHAVGKLIER
MAB_0418|M.abscessus_ATCC_1997 KTANVILGNAFDIPGITVDTHFGRLVRRWRWTEEEDPVKVEHLVGELIER
*****:*****.:*********.**.:***** .****:* *..****
MMAR_5162|M.marinum_M KEWTLLSHRVIFHGRRVCHARKPACGVCVLAKDCPSYGLGPTEPLLAAPL
MUL_4248|M.ulcerans_Agy99 KEWTLLSHRVIFHGRRVCHARKPACGVCVLAKDCPSYGLGPTEPLLAAPL
Mb3698c|M.bovis_AF2122/97 KEWTLLSHRVIFHGRRVCHARRPACGVCVLAKDCPSFGLGPTEPLLAAPL
Rv3674c|M.tuberculosis_H37Rv KEWTLLSHRVIFHGRRVCHARRPACGVCVLAKDCPSFGLGPTEPLLAAPL
MLBr_02301|M.leprae_Br4923 DQWTLLSHRVIFHGRRVCHARKPACGVCVLAKDCPSFGLGPTEPLLAAPL
MAV_0455|M.avium_104 SEWTMLSHRVIFHGRRVCHSRKPACGVCLLARDCPSFGLGPTEPPLAAQL
Mflv_1373|M.gilvum_PYR-GCK SEWTLLSHRVIFHGRRVCHARKPACGVCVLAKDCPSYGTGPTDPLIAAAL
Mvan_5433|M.vanbaalenii_PYR-1 SEWTLLSHRVIFHGRRVCHARKPACGVCVLAKDCPSYGAGPTDPLIAAPL
MSMEG_6187|M.smegmatis_MC2_155 SEWTLLSHRVIFHGRRVCHARKPACGVCVLAKDCPSYGIGPTDPPTAAAL
TH_2130|M.thermoresistible__bu REWTLLSHRVIFHGRRVCHARKPACGVCVLADDCPSYGIGETDKLAAAEL
MAB_0418|M.abscessus_ATCC_1997 KEWTLLSHRVIFHGRRVCHARKPACGVCVLAKDCPSYGLGPTDPEVAAPL
:**:**************:*:******:** ****:* * *: ** *
MMAR_5162|M.marinum_M VQGPETEHLLAMAGL
MUL_4248|M.ulcerans_Agy99 VQGPETEHLLAMAGL
Mb3698c|M.bovis_AF2122/97 VQGPETDHLLALAGL
Rv3674c|M.tuberculosis_H37Rv VQGPETDHLLALAGL
MLBr_02301|M.leprae_Br4923 VQGPEAGHLLALAGL
MAV_0455|M.avium_104 VRGPETEHLLALAGL
Mflv_1373|M.gilvum_PYR-GCK VKGPETEHLLALAGL
Mvan_5433|M.vanbaalenii_PYR-1 VKGPETEHLLALAGL
MSMEG_6187|M.smegmatis_MC2_155 VKGPETEHLLALAGL
TH_2130|M.thermoresistible__bu VRGPERDHLLALAGL
MAB_0418|M.abscessus_ATCC_1997 VKGPETEHLLALAGL
*:*** ****:***