For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MATTATEADTGRTLSGHDEAAEHIARQAFADAQVGAVGLELESHTVELTAPHRRITWNRLREVGDSVPDL PGSSAITFEPGGAVELSGPPCSDVWSAISSMRADHEILTAVYRGAGIALASLGTDPLRRPERVNPGARYV AMERHFGAAGYGETALQMMTCTASLQVNVQSGTPRQWRDRFVLAQRIGPTMAALSASSPMLTGRPTGRRN TRQWIWDNLDPRRCAPVEIGVDPTESWVEYALRAPVMLVRNKDGADAVVTRVPFQSWVDGTMPLAGRAPT TEDLDYHLTTLFPPVRPRRWLELRYLDAAPDWWWPALAFTAVAALDDPQVADIAAEIAEPVGNAWGVAAR IGLEDPALHAAAHRLVSAACAVAPPELATDMEFLLERVEQGRCPADDFIDNVTEYGVEKAFSGAIG
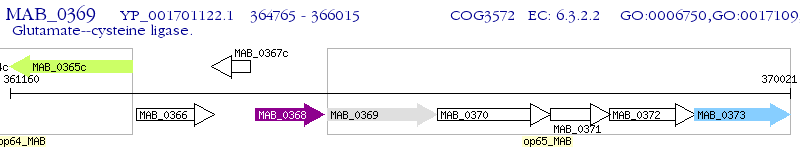
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0369 | - | - | 100% (416) | glutamate--cysteine ligase GshA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3730c | gshA | 1e-107 | 49.75% (398) | glutamate--cysteine ligase gshA (gamma-glutamylcysteine |
| M. gilvum PYR-GCK | Mflv_1315 | - | 1e-109 | 51.25% (400) | glutamate--cysteine ligase, GCS2 |
| M. tuberculosis H37Rv | Rv3704c | gshA | 1e-107 | 49.75% (398) | glutamate--cysteine ligase gshA (gamma-glutamylcysteine |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5215 | gshA | 1e-113 | 50.00% (414) | glutamate--cysteine ligase GshA |
| M. avium 104 | MAV_0399 | - | 1e-115 | 53.28% (396) | putative glutamate--cysteine ligase |
| M. smegmatis MC2 155 | MSMEG_6250 | - | 1e-108 | 52.02% (396) | glutamate--cysteine ligase |
| M. thermoresistible (build 8) | TH_0772 | gshA | 1e-109 | 50.24% (414) | GLUTAMATE--CYSTEINE LIGASE GSHA (GAMMA-GLUTAMYLCYSTEINE |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5486 | - | 1e-110 | 51.88% (399) | glutamate--cysteine ligase, GCS2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3730c|M.bovis_AF2122/97 ---------------------MTLAAMTAAASQLDNAAPDD----VEITD
Rv3704c|M.tuberculosis_H37Rv ---------------------MTLAAMTAAASQLDNAAPDD----VEITD
MMAR_5215|M.marinum_M ---------------------MTFAPSPAATSQLDSSRAAD----ADIAG
MAV_0399|M.avium_104 ---------------------MTFAPSTAAASRPAGGDPAAELRIAELTD
Mflv_1315|M.gilvum_PYR-GCK MNLSVLSGRWVKTQRPFHTRRGWLMAHTMASDGVVEHGVVP------LTS
Mvan_5486|M.vanbaalenii_PYR-1 ------------------------MACTMTSDGVVEQRFDP------LSS
MSMEG_6250|M.smegmatis_MC2_155 ------------------------MALPARSDSGCAVPVE-------FTS
TH_0772|M.thermoresistible__bu ------------------------MTLPVTAEAPAT-----------VDS
MAB_0369|M.abscessus_ATCC_1997 ------------------------MATTATEADTGRT----------LSG
. . . .
Mb3730c|M.bovis_AF2122/97 SSAAAEYIADGCLVDGPLGRVGLEMEAHCFDPADPFRRPSWEEITEVLEW
Rv3704c|M.tuberculosis_H37Rv SSAAAEYIADGCLVDGPLGRVGLEMEAHCFDPADPFRRPSWEEITEVLEW
MMAR_5215|M.marinum_M SAVAAQYIANGCLVDGPIGRVGLEIEAHCYDPADPHRRPSWEEIGDVLEG
MAV_0399|M.avium_104 SAAVAHYIARGCLADAPLGRVGLEVEAHCHDPQDPCRRPGWDEIAAVLDA
Mflv_1315|M.gilvum_PYR-GCK AEDAAQHIVDTCLKDVPPGPVGLEIEAHCYDLSDPFRRPGWDELTAVIAD
Mvan_5486|M.vanbaalenii_PYR-1 AEAAAQHIVDGCLIDGPVGQVGLEVEAHCFDLTDPSRRPGWDELTATITA
MSMEG_6250|M.smegmatis_MC2_155 AEQAAAHIGANSLQDGPIGRVGLEIEAHCFDLSNPTRRPSWDELSAVIAD
TH_0772|M.thermoresistible__bu ARRAAAHIEASCLRDGPVGRVGLEIEAHCFDVADPGRRPGWAELTDVIAT
MAB_0369|M.abscessus_ATCC_1997 HDEAAEHIARQAFADAQVGAVGLELESHTVELTAPHRRITWNRLREVGDS
.* :* .: * * ****:*:* : * ** * .: .
Mb3730c|M.bovis_AF2122/97 LSPLPGGSVVSVEPGGAVELSGPPADGVLAAIGAMTRDQAVLRSALANAG
Rv3704c|M.tuberculosis_H37Rv LSPLPGGSVVSVEPGGAVELSGPPADGVLAAIGAMTRDQAVLRSALANAG
MMAR_5215|M.marinum_M LGKPPGGSAVTVEPGGAVELSGPPCDGILAAIDAMHRDQAVLRSAFADAG
MAV_0399|M.avium_104 LPALPGGSRVTVEPGGAVELSGPPADGVLAAIDAMRRDQAVLRPAFAEAG
Mflv_1315|M.gilvum_PYR-GCK LPRLPGGSLVTVEPGGAVELSGPPATGTADAVSAMVTDRAVLRDVFAGAG
Mvan_5486|M.vanbaalenii_PYR-1 LPPLPGGSVVTVEPGGAVELSGPPLAEPASAISAMAADHTVLRAAFARAG
MSMEG_6250|M.smegmatis_MC2_155 VPPLPGGSRITVEPGGAVELSGPPYDGPLAAVAALQADRAVLRAEFARRN
TH_0772|M.thermoresistible__bu VGELPGGSAISVEPGGAVELSGPPLDGPVAAATAMLTDQAVLRAAFARAG
MAB_0369|M.abscessus_ATCC_1997 VPDLPGSSAITFEPGGAVELSGPPCSDVWSAISSMRADHEILTAVYRGAG
: **.* ::.************ * :: *: :* .
Mb3730c|M.bovis_AF2122/97 LGLVFLGADPLRSPVRVNPGARYRAMEQFFAASHSGVPGAAMMTSTAAIQ
Rv3704c|M.tuberculosis_H37Rv LGLVFLGADPLRSPVRVNPGARYRAMEQFFAASHSGVPGAAMMTSTAAIQ
MMAR_5215|M.marinum_M LGLVCLGADPIRPTMRINPGARYQAMEQFFTASQSGMAGAAMMTSTASIQ
MAV_0399|M.avium_104 LGLVFLGADPLRPPQRINPGARYRAMERFFATSRSGEAGAAMMTSTASIQ
Mflv_1315|M.gilvum_PYR-GCK LGLVLLGADPLRPAGRVNPGSRYAAMERFFAESGTPEAGAAMMTSTASVQ
Mvan_5486|M.vanbaalenii_PYR-1 LGLVLVGADPLRPAARVNPGPRYQAMEAFFRASDSGDAGAAMMTSTASVQ
MSMEG_6250|M.smegmatis_MC2_155 LGLVLLGTDPLRPTRRVNPGARYSAMEQFFTASGTAEAGAAMMTATASVQ
TH_0772|M.thermoresistible__bu YALVMLGADPVRQPQRVNPGSRYRAMETFFAASGTAAAGSAMMTSTASIQ
MAB_0369|M.abscessus_ATCC_1997 IALASLGTDPLRRPERVNPGARYVAMERHFGAAGYGETALQMMTCTASLQ
.*. :*:**:* . *:***.** *** .* : .. ***.**::*
Mb3730c|M.bovis_AF2122/97 VNLDAGPQEGWAERVRLAHALGPTMIAIAANSPMLGGRFSGWQSTRQRVW
Rv3704c|M.tuberculosis_H37Rv VNLDAGPQEGWAERVRLAHALGPTMIAIAANSPMLGGRFSGWQSTRQRVW
MMAR_5215|M.marinum_M VNLDAGPQSGWASRVRLAHALGPTMIAIAANSPMLGGEFSGWRSTRQRVW
MAV_0399|M.avium_104 VNVDAGPREGWAARVRLAHALGPTMIAMTANSPMLDGDFTGWVSSRQRVW
Mflv_1315|M.gilvum_PYR-GCK VNLEAGHRAGWADRVRLAHALGPTMIAIAANSPVLAGKFTGWHSTRQLVW
Mvan_5486|M.vanbaalenii_PYR-1 VNLEAGPRAGWADRVRLAHALGPTMIAIAANSPVLGGDFTGWQSTRQLVW
MSMEG_6250|M.smegmatis_MC2_155 VNLDAGPRDGWAERVRLAHALGPTMIAITANSPMLGGQFTGWCSTRQRVW
TH_0772|M.thermoresistible__bu VNVEAGPRTGWAERIRLAHALGPVMVAIAANSPLLGGRFSGWRSSRQRIW
MAB_0369|M.abscessus_ATCC_1997 VNVQSGTPRQWRDRFVLAQRIGPTMAALSASSPMLTGRPTGRRNTRQWIW
**:::* * *. **: :**.* *::*.**:* * :* .:** :*
Mb3730c|M.bovis_AF2122/97 GQMDSARCGPILG-ASGD-HPGIDWAKYALKAPVMMVRSPDTQDTRAVTD
Rv3704c|M.tuberculosis_H37Rv GQMDSARCGPILG-ASGD-HPGIDWAKYALKAPVMMVRSPDTQDTRAVTD
MMAR_5215|M.marinum_M GQMDDGRCGPILG-ASAD-DPATDWAHYALKAPVMLVHNP---DAVAVTH
MAV_0399|M.avium_104 GRMDSARCGPILG-ASGD-DPGTDWARYALKAPVMLVHTP---DAEAVTR
Mflv_1315|M.gilvum_PYR-GCK SQLDSARCGPILG-VDGD-DPASDWARYALRAPVMLVHTP---EAVPVTQ
Mvan_5486|M.vanbaalenii_PYR-1 SRLDSARCGPILG-ASGD-DPASDWARYALRAPVMLVHSP---DPTPVTQ
MSMEG_6250|M.smegmatis_MC2_155 GQLDSARCGPVLG-VDGD-DPASEWARYALRAPVMLVNSP---DAVPVTN
TH_0772|M.thermoresistible__bu GQLDSARCGPILGGRDGVRTPGGDWARYALRAPVMLVNGAGDRTAEPVTD
MAB_0369|M.abscessus_ATCC_1997 DNLDPRRCAPVEI----GVDPTESWVEYALRAPVMLVRNKD--GADAVVT
..:* **.*: * .*..***:****:*. . .*.
Mb3730c|M.bovis_AF2122/97 YVPFTDWVDGRVLLDGRRATVADLVYHLTTLFPPVRPRQWLEIRYLDSVP
Rv3704c|M.tuberculosis_H37Rv YVPFTDWVDGRVLLDGRRATVADLVYHLTTLFPPVRPRQWLEIRYLDSVP
MMAR_5215|M.marinum_M HVPFADWVDGRVLLGDRRPTYADLDYHLTTLFPPVRPRQWLEIRYLDSLS
MAV_0399|M.avium_104 WVPFADWVDGRVLLGGRRPTVADLEYHLTTLFPPVRPRQWLEIRYLDSVP
Mflv_1315|M.gilvum_PYR-GCK WVPFADWADGRVLLDDRRPTLADLDYHLTTLFPPVRPRGFLEVRYLDSVP
Mvan_5486|M.vanbaalenii_PYR-1 WVPFADWADGRVLLDDRPPTLADLDYHLTTLFPPVRPRGFLEVRYLDSVP
MSMEG_6250|M.smegmatis_MC2_155 WVPFADWADGRAVLGGRRPTEADLDYHLTTLFPPVRPRRWLEIRYLDSVP
TH_0772|M.thermoresistible__bu WVPFADWADGRTALGGRRPTVADLEYHLTTLFPPVRPRGWLEIRYLDCVP
MAB_0369|M.abscessus_ATCC_1997 RVPFQSWVDGTMPLAGRAPTTEDLDYHLTTLFPPVRPRRWLELRYLDAAP
*** .*.** * .* .* ** ************* :**:****. .
Mb3730c|M.bovis_AF2122/97 DEVWPAVVFTLVTLLDDPVAADLAVDAVEPVATAWDTAARIGLADRRLYL
Rv3704c|M.tuberculosis_H37Rv DEVWPAVVFTLVTLLDDPVAADLAVDAVEPVATAWDTAARIGLADRRLYL
MMAR_5215|M.marinum_M DVFWPAVVFTLVTLLDDPGAAELAFEAVQPVATEWDTAARAGLGDPRLHA
MAV_0399|M.avium_104 DTLWPAVVFTLVALLDDPVAAGLAARAVEPVATAWDVAARVGLRDRRLYA
Mflv_1315|M.gilvum_PYR-GCK DAVWPAVVFTLSTLLDDPVAADVAAEATEPVATSWDRAAQLGLDDRRLHT
Mvan_5486|M.vanbaalenii_PYR-1 DATWPAVVYTLATLLDDPAAADVAAEATEEVATAWDRAAQLGLRDRRLHA
MSMEG_6250|M.smegmatis_MC2_155 DALWPAAVFTLTTLLDDPVAAESAAEATRPVATAWDRAARMGLTDRHLHT
TH_0772|M.thermoresistible__bu DALWPAVVFTLVTLLDDPSAADIAAEATEPVSSAWSRAARDGLADRRLFT
MAB_0369|M.abscessus_ATCC_1997 DWWWPALAFTAVAALDDPQVADIAAEIAEPVGNAWGVAARIGLEDPALHA
* *** .:* : **** .* * .. *.. *. **: ** * *.
Mb3730c|M.bovis_AF2122/97 AANRCLAIAARRVPTELIGAMQRLVDHVDRGVCPADDFSDRVIAGGIASA
Rv3704c|M.tuberculosis_H37Rv AANRCLAIAARRVPTELIGAMQRLVDHVDRGVCPADDFSDRVIAGGIASA
MMAR_5215|M.marinum_M AASRCVAIAAERAPAELADALQSLLAYVQRGRCPGDDFGDQVTEHGVAPA
MAV_0399|M.avium_104 AANACLAIAAERAPAELAEPMRRLVRLVEQGRCPGDDFADQVIEHGIAAS
Mflv_1315|M.gilvum_PYR-GCK AAVRCVYAAADRAPAALVESMELLTRSVEQGRSPGDDFADRVVNDGIAAA
Mvan_5486|M.vanbaalenii_PYR-1 AATRCVQAAADRAPRALEESMGHLVRSVEQGRSPADDFADRVVAHGIADA
MSMEG_6250|M.smegmatis_MC2_155 AALTCVRLAAERAPAELEESMTLLMRSVQQRRSPADDFSDRVVARGIAAA
TH_0772|M.thermoresistible__bu AANRCVRTAAERTPAPLLESMERLVRSVERGRCPADDFADLVVRRGIGDA
MAB_0369|M.abscessus_ATCC_1997 AAHRLVSAACAVAPPELATDMEFLLERVEQGRCPADDFIDNVTEYGVEKA
** : *. .* * : * *:: .*.*** * * *: :
Mb3730c|M.bovis_AF2122/97 VTGMMHGAS---
Rv3704c|M.tuberculosis_H37Rv VTGMMHGAS---
MMAR_5215|M.marinum_M IARLARGEQ---
MAV_0399|M.avium_104 VSRAARGPQGGP
Mflv_1315|M.gilvum_PYR-GCK VTRLAQGDM---
Mvan_5486|M.vanbaalenii_PYR-1 VTELAQGEL---
MSMEG_6250|M.smegmatis_MC2_155 VRELAKGEL---
TH_0772|M.thermoresistible__bu VDRHGRGDW---
MAB_0369|M.abscessus_ATCC_1997 FSGAIG------
.