For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVRIMSWRTAGFWLAAGLVAVLLQHWLIPLNAPNSFGLFSNGGDLMTYRFGGLRVLHRELLYATEIPDA GWFTYTPFAAMLFAPLGFVTLGAAKVIWFLVNLGALFIIIWRCWRVLGFSANAPLAIACVGMNLVAWDIQ PIHGTLWQGQVNLVLAALIIWDLTRPEGARWRGWSVGLASGVKLTAVIFVPYLLITRQWRAVAAAAVTAI GTVVLGWIVLPSDSSDYWLGAVRTTGHIGSLDHPANSSIGGALANVYTPQPMPTWLWVLLAGGAALLGMA AAWRAHRTGRELLAITVVGMVGCVVSPLAWTHHWVWTVPLMVLLVNQIMTTQGGARWRWAAATTVIAVLV SMWWYWGLYVRAMRLNPDFESYITGWGAVIAHMSRAERVFDTALYPLLFLTVATWVLATKSLRAGTVQE
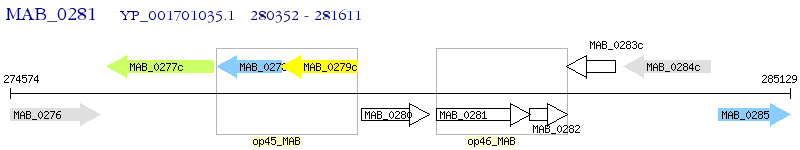
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0281 | - | - | 100% (419) | hypothetical protein MAB_0281 |
| M. abscessus ATCC 19977 | MAB_0096 | - | 2e-75 | 37.78% (405) | hypothetical protein MAB_0096 |
| M. abscessus ATCC 19977 | MAB_1985 | - | 2e-46 | 34.72% (360) | hypothetical protein MAB_1985 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1190 | pimE | 1e-36 | 32.51% (323) | mannosyltransferase |
| M. gilvum PYR-GCK | Mflv_0993 | - | 9e-64 | 40.06% (322) | hypothetical protein Mflv_0993 |
| M. tuberculosis H37Rv | Rv1159 | pimE | 1e-36 | 32.51% (323) | mannosyltransferase |
| M. leprae Br4923 | MLBr_00893 | - | 5e-32 | 30.06% (346) | hypothetical protein MLBr_00893 |
| M. marinum M | MMAR_4292 | pimE | 8e-35 | 31.25% (320) | transmembrane protein |
| M. avium 104 | MAV_1298 | pimE | 2e-35 | 31.58% (342) | mannosyltransferase |
| M. smegmatis MC2 155 | MSMEG_5149 | pimE | 1e-35 | 31.36% (338) | mannosyltransferase |
| M. thermoresistible (build 8) | TH_1105 | - | 2e-33 | 33.03% (330) | Probable conserved integral membrane protein |
| M. ulcerans Agy99 | MUL_1006 | pimE | 8e-34 | 30.63% (320) | mannosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_3543 | - | 8e-35 | 33.23% (334) | putative integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1190|M.bovis_AF2122/97 MCRTLIDGPVRSAIAKVRQIDTTSSTPAAAR---RVTSPPARETRAAVLL
Rv1159|M.tuberculosis_H37Rv MCRTLIDGPVRSAIAKVRQIDTTSSTPAAAR---RVTSPPARETRAAVLL
MMAR_4292|M.marinum_M MTGHLGN----WVGTRVGRIDTTSAAPAASTTRHKAVSPKVWGRRGAPLL
MUL_1006|M.ulcerans_Agy99 MTGHLGN----WVGTRVGRIDTTSAAPAASTTRHKAVSPKVWGRRGAPLL
MAV_1298|M.avium_104 --------------------------------------------MAAPLL
MSMEG_5149|M.smegmatis_MC2_155 ------------------------------------------------ML
TH_1105|M.thermoresistible__bu ----------------------------------------------MFQL
Mvan_3543|M.vanbaalenii_PYR-1 --------------------------MSNRQSPGGAGWKPTLA-WRLFQL
MLBr_00893|M.leprae_Br4923 --------------------------MSRQQTPEVGNKHRQIGRCCLLWL
MAB_0281|M.abscessus_ATCC_1997 ------------------------MTVRIMSWRTAGFWLAAGLVAVLLQH
Mflv_0993|M.gilvum_PYR-GCK --------------------------MSRLALSRPALGVLAGWASLSVVL
Mb1190|M.bovis_AF2122/97 LVLSVGARLAWTYLAPNGANFVDLHVY-VSGAASLDHPGTLYGYVYADQT
Rv1159|M.tuberculosis_H37Rv LVLSVGARLAWTYLAPNGANFVDLHVY-VSGAASLDHPGTLYGYVYADQT
MMAR_4292|M.marinum_M LVFSVAARLAWTYLTSNGANFVDLHVY-VGGAAALDHPGSLYSYVYADQT
MUL_1006|M.ulcerans_Agy99 LVFSVAARLAWTYLTSNGANFVDLHVY-VGGAAALGHPGSLYSYVYADQT
MAV_1298|M.avium_104 LVASVAARLAWTYLVPNGANFVDLHVY-LGGAAALDHPGTLYSYVYADQT
MSMEG_5149|M.smegmatis_MC2_155 LVVSILARLAWTYLVPNGANFVDLHVY-VGGADALDGPGALYDYVYADQT
TH_1105|M.thermoresistible__bu ITLAALIWAGWRLLG-HLPYRIDIDVYRMGGRAWLDGDPLYADGAIFETR
Mvan_3543|M.vanbaalenii_PYR-1 LTFAALAAAAWRLLG-HVPYRIDIDVYRMGGRAWLDGQSLYADGAMFQTR
MLBr_00893|M.leprae_Br4923 LAVTALGQVAWQLLG-HTPYRIDIDIYQMGGQAWLDGRPLYSGDVLFHTP
MAB_0281|M.abscessus_ATCC_1997 WLIPLNAPNSFGLFS----NGGDLMTYRFGGLRVLHR------ELLYATE
Mflv_0993|M.gilvum_PYR-GCK ALTTVGPTANWGLLD----GGGDLDVYRRGAWHVLSG------LPLYDFP
. : : *: * .. *
Mb1190|M.bovis_AF2122/97 PDFPLPFTYPPFAAVVFYPLHLVPFGLIALLWQVVTMAALYGAVRISQRL
Rv1159|M.tuberculosis_H37Rv PDFPLPFTYPPFAAVVFYPLHLVPFGLIALLWQVVTMAALYGAVRISQRL
MMAR_4292|M.marinum_M PDFPLPFTYPPFAAVVFYPLHLLPFGLVAFLWQLATIAALYGAVRISQRL
MUL_1006|M.ulcerans_Agy99 PDFPLPFTYPPFAAVVFYPLHLLPFGLVAFLWQLATITALYGAVRISQRL
MAV_1298|M.avium_104 PDFPLPFTYPPFAAIVFYPLHLLPFGLVALGWQLATIAALYGAVRISQRL
MSMEG_5149|M.smegmatis_MC2_155 PDFPLPFTYPPFAAIVFYPLHLLPFGVVAFIWQIGIIAALYGVVRVSQRL
TH_1105|M.thermoresistible__bu AGLNLPFTYPPLAAVAFAPFAWLSLEAAAVAITLTTLVLLVVATVIVLTR
Mvan_3543|M.vanbaalenii_PYR-1 GGLELPFTYPPLAAVAFAPFALLSLEAASVAITLTTLILLLVSTVIVLTR
MLBr_00893|M.leprae_Br4923 IGLNLPFTYPPLAAIAFCPFAWLHMPAASVAITLTTLMLLIVSTIIVLTR
MAB_0281|M.abscessus_ATCC_1997 IPDAGWFTYTPFAAMLFAPLGFVTLGAAKVIWFLVNLGALFIIIWRCWRV
Mflv_0993|M.gilvum_PYR-GCK VDKKLFYTYTPFSALVFAPLELLPAEPDRHIWLALNLAVLIAVVVQSWRL
:**.*::*: * *: : : *
Mb1190|M.bovis_AF2122/97 MGGTA-----------ETGHFAAMLWTAIAIWIEPLRSTFDYGQINVLLM
Rv1159|M.tuberculosis_H37Rv MGGTA-----------ETGHFAAMLWTAIAIWIEPLRSTFDYGQINVLLM
MMAR_4292|M.marinum_M IGVPA-----------ERGHRAAMVWTAVAIWIEPLRSTFDYGQINVLLM
MUL_1006|M.ulcerans_Agy99 IGVPA-----------ERGHRAAMVWTAVAIWIEPLRSTFDYGQINVLLM
MAV_1298|M.avium_104 LGVAAG----------AHGRRVAMVWTAVTIWIEPLRSTFDYGQVNVLLM
MSMEG_5149|M.smegmatis_MC2_155 MGLQS-------------QRRVAMLWTALGIWTEPLRSTFDYGQVNVVLV
TH_1105|M.thermoresistible__bu LDVWPDTAMTTEPAWLRRSWLAAAIVAPAIILLEPIRSNFEFGQINVVLM
Mvan_3543|M.vanbaalenii_PYR-1 LDVWPSTRVTSEPAWLRRCWLAAAIVAPAVLYLEPLRANFDFGQINVVLM
MLBr_00893|M.leprae_Br4923 LDVWAMSRAVPGPAWLRRLWLAVIIVSPAAIWLEPISSNFAFGQINVVLM
MAB_0281|M.abscessus_ATCC_1997 LGFSAN----------APLAIACVGMNLVAWDIQPIHGTLWQGQVNLVLA
Mflv_0993|M.gilvum_PYR-GCK LGYRVD----------RALVGVSVLMALGCVFVEPVRTTLFFGQINLILM
:. . :*: .: **:*::*
Mb1190|M.bovis_AF2122/97 LAALWAVYTPR-WWLSGLLVGVASGVKLTPAITAVYLVGVRRLHAAAFSV
Rv1159|M.tuberculosis_H37Rv LAALWAVYTPR-WWLSGLLVGVASGVKLTPAITAVYLVGVRRLHAAAFSV
MMAR_4292|M.marinum_M LAALWAVYTTR-WWLSGLLVGVAAGIKLTPAITAVYLAGVRRWRAAVCSA
MUL_1006|M.ulcerans_Agy99 LAALWAVYTTR-WWLSGLLVGVAAGIKLTPAITAVYLAGVRRWRAAVCSA
MAV_1298|M.avium_104 TAVLWAVYSTR-WWVSALLVGLAAGIKLTPAVSGVYLAGARRWAATVFSA
MSMEG_5149|M.smegmatis_MC2_155 LAVLCAVSTTR-WWLSGLLVGLAAGIKLTPAVAGLYFLGARRWAAVACSA
TH_1105|M.thermoresistible__bu TLVIADCVPRRTPWPRGALLGVAIALKLTPAVFLLYFLLRRDGRALITAS
Mvan_3543|M.vanbaalenii_PYR-1 TLVIADCVPRRTPWPRGVLLGVAIAVKLTPAVFLLYFLLRRDVRALLVTA
MLBr_00893|M.leprae_Br4923 TLVIADCVPRRTPWPRGLMLGLGIALKLTPAVFLLYFVLRRDNRAALTSW
MAB_0281|M.abscessus_ATCC_1997 ALIIWDLTRPEGARWRGWSVGLASGVKLTAVIFVPYLLITRQWRAVAAAA
Mflv_0993|M.gilvum_PYR-GCK LLILSDTVGATRSRLAGIGTGLAAGIKLTPLYFVLYFLTLRQRRVAVTAV
: . *:. .:***. *: * . :
Mb1190|M.bovis_AF2122/97 VVFLATVGVSLLVVGDEARYYFTDLLGDAGRVGP-IATSFNQSWRGAISR
Rv1159|M.tuberculosis_H37Rv VVFLATVGVSLLVVGDEARYYFTDLLGDAGRVGP-IATSFNQSWRGAISR
MMAR_4292|M.marinum_M IVFLVSVGVSLLIVGGQARYYFTDLLGDAHRVGP-IATSFNQSWRGAISR
MUL_1006|M.ulcerans_Agy99 IVFLVSVGVSLLIVGGQARYYFTDLLGDAHRVGP-IATSFSQSWRGAISR
MAV_1298|M.avium_104 VIFLATIGISVLVVGDQTRYYFTELLGDAHRVGP-IATSFNQSWRGGLSR
MSMEG_5149|M.smegmatis_MC2_155 AVFFATVGVSWLVVGAQARRYFTELLGDADRIGP-IGTSFNQSWRGGISR
TH_1105|M.thermoresistible__bu VTAVLATLAGFALAWRDSWEYWTHTVSNTDRIGT-ATLNTNQNIAGALAR
Mvan_3543|M.vanbaalenii_PYR-1 ASAVVATLAGFALAWRDSWEYWLETVRNTDRIGT-ATLNTNQNIAGVLAR
MLBr_00893|M.leprae_Br4923 ASFAGATLVGFVLAWRDSWEYWTHTVHNTERIGA-AALNTDQNIAGALAR
MAB_0281|M.abscessus_ATCC_1997 VTAIGTVVLGWIVLPSDSSDYWLGAVRTTGHIGS-LDHPANSSIGGALAN
Mflv_0993|M.gilvum_PYR-GCK AVFAATVVVGLILLPRDSLTYWAGTFLRSERIGDRLGHPSNQSLRGMLAR
: . : :: *: . : ::* ... * ::.
Mb1190|M.bovis_AF2122/97 I-LGHDAGFGPLVLAAIASTAVLAILAWRALD-----RSDRLGKLLVVEL
Rv1159|M.tuberculosis_H37Rv I-LGHDAGFGPLVLAAIASTAVLAILAWRALD-----RSDRLGKLLVVEL
MMAR_4292|M.marinum_M I-LGHDAGFGPLVLAAIAVTAVLAVLAWRALE-----PADHLGKLLVVEL
MUL_1006|M.ulcerans_Agy99 I-LGHDAGFGPLVLAAIAVTAVLAVLAWRALE-----PADHLGKLLVVEL
MAV_1298|M.avium_104 I-LGHDVGFGPLLVAAVAGTAVLAVLAWRALD-----RSDRLGRLIVVEV
MSMEG_5149|M.smegmatis_MC2_155 I-LGHDAGFGPLVLIGIGITAVLALLAWRAIGG----AQDRLGGILVVSL
TH_1105|M.thermoresistible__bu LGLDEDTRFLLWVLACFAVLALTVWAVRRVLRSRVCADEASVLALICVAM
Mvan_3543|M.vanbaalenii_PYR-1 LGLGEGERFVVWTVLCFAVLGLTVWAARRALR-----AEQPVLALICVAM
MLBr_00893|M.leprae_Br4923 LGLGEHERFLLWAAGCLLVLAATAWAMLRVLR-----AGEPMLAVICVAL
MAB_0281|M.abscessus_ATCC_1997 VYTPQPMPTWLWVLLAGGAALLGMAAAWRAHR-----TGRELLAITVVGM
Mflv_0993|M.gilvum_PYR-GCK LLDDEP-PVWLWLALAVLIGVASLTLAVWLHR-----TGERLLSVTVVGL
: . : : * :
Mb1190|M.bovis_AF2122/97 FGLLLSPISWTHHWVWLVPLMIWLIDGPAR-----------ERPGARILG
Rv1159|M.tuberculosis_H37Rv FGLLLSPISWTHHWVWLVPLMIWLIDGPAR-----------ERPGARILG
MMAR_4292|M.marinum_M FGLLLSPISWTHHWVWLVPLMIWLLSGPLR-----------DRLGARILG
MUL_1006|M.ulcerans_Agy99 FGLLLSPISWTHHWVWLVPLMIWVLSGPLR-----------DRLGARILG
MAV_1298|M.avium_104 FGLLLSPISWTHHWVWLVPLMIWAIHGPVR-----------AWRGARLVG
MSMEG_5149|M.smegmatis_MC2_155 FGLVLSPISWTHHWVWLIPLMMWLLHGPLS-----------ALRGARILG
TH_1105|M.thermoresistible__bu FGLVVSPVSWSHHWVWALPAVLVAAVVSARHR-------VRALGVTAAAG
Mvan_3543|M.vanbaalenii_PYR-1 FGLVVSPVSWSHHWVWALPTVLVTAVLAVRLR-------HVALAVVSAAG
MLBr_00893|M.leprae_Br4923 FGLVVSPVSWSHHWVWMLPAVLVTGILSWRRQ-------NVAVAVISAAG
MAB_0281|M.abscessus_ATCC_1997 VGCVVSPLAWTHHWVWTVPLMVLLVNQIMTTQGGARWRWAAATTVIAVLV
Mflv_0993|M.gilvum_PYR-GCK SSAVVSPFSWTHHWVWVVPLMVWFVHRALT-----CWRWWLAPAALFVAL
. ::**.:*:***** :* ::
Mb1190|M.bovis_AF2122/97 WGWLVLTIVGVPWLLSFAQPSIW-QIGRPWYLAWAGLVYVVATLATLGWI
Rv1159|M.tuberculosis_H37Rv WGWLVLTIVGVPWLLSFAQPSIW-QIGRPWYLAWAGLVYVVATLATLGWI
MMAR_4292|M.marinum_M WGWLALTVIGVPWLLSFAQPTIW-QIGRPWYLAWAGLVYVVATLATLGWI
MUL_1006|M.ulcerans_Agy99 WGWLALTVIGVPWLLSFAQPTIW-QIGRPWYLAWAGLVYVVATLATLGWI
MAV_1298|M.avium_104 WGWLALTVIGVPWLLSFAQPNIW-QIGRPWYLAWAGLVYIVAAVATLVWI
MSMEG_5149|M.smegmatis_MC2_155 WGWLALTLLGVPWLLSFAQPTIW-EIGRPWYLAWAGLVYIVATLATLGWI
TH_1105|M.thermoresistible__bu VALLVWTPIRLLPENEEMTASLWRQLLGGSYVWWALVLIVVIGALGS-GS
Mvan_3543|M.vanbaalenii_PYR-1 LALMVWTPITLLPEHRETTASLWRQLVGGSYLWWALAVIVVSATVTLPRP
MLBr_00893|M.leprae_Br4923 VALMRWTPIGLLPQHQETTAVWWRQLAGMSYVWWALAVIVATGLTVTTRV
MAB_0281|M.abscessus_ATCC_1997 SMWWYWGLYVRAMRLNPDFESYITGWGAVIAHMSRAERVFDTALYPLLFL
Mflv_0993|M.gilvum_PYR-GCK GAWPFWFRNLADPRIGFYLFPYTH---VPEVLLRNAYLWLFAVLLIWLGV
.
Mb1190|M.bovis_AF2122/97 AASERYVRIRPRRMAN-----
Rv1159|M.tuberculosis_H37Rv AASERYVRIRPRRMAN-----
MMAR_4292|M.marinum_M ATTRQRAR-------------
MUL_1006|M.ulcerans_Agy99 ATTRQRAR-------------
MAV_1298|M.avium_104 AAAGRRAGHQVTAPSVTPA--
MSMEG_5149|M.smegmatis_MC2_155 AFSRKGSG-------------
TH_1105|M.thermoresistible__bu ARPRP-VDAERVPTGG-----
Mvan_3543|M.vanbaalenii_PYR-1 GRDAPGLGAAQPATNGVKA--
MLBr_00893|M.leprae_Br4923 TSQDSATQTLLTPVSTAS---
MAB_0281|M.abscessus_ATCC_1997 TVATWVLATKSLRAGTVQE--
Mflv_0993|M.gilvum_PYR-GCK RAARSVAGAKAAGEEPVRAEE