For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAPPVHDRAHHPVRDVIVIGSGPAGYTAALYAARAQLAPLVFEGTSFGGALMTTTDVENYPGFRNGITG PELMDEMREQALRFGADLRMEDVESVSLHGPLKSVVTADGQTHRARAVILAMGAAARYLQVPGEQELLGR GVSSCATCDGFFFRDQDIAVIGGGDSAMEEATFLTRFARSVTLVHRRDEFRASKIMLDRARNNDKIRFLT NHTVVAVDGDTTVTGLRVRDTNTGAETTLPVTGVFVAIGHEPRSGLVREAIDVDPDGYVLVQGRTTSTSL PGVFAAGDLVDRTYRQAVTAAGSGCAAAIDAERWLAEHAATGEADSTDALIGAQR
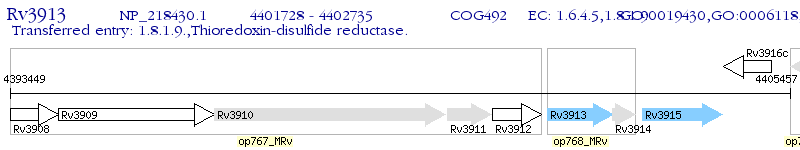
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3913 | trxB2 | - | 100% (335) | PROBABLE THIOREDOXIN REDUCTASE TRXB2 (TRXR) (TR) |
| M. tuberculosis H37Rv | Rv1869c | - | 2e-08 | 24.66% (292) | reductase |
| M. tuberculosis H37Rv | Rv0252 | nirB | 1e-07 | 24.93% (349) | nitrite reductase large subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3944 | trxB2 | 0.0 | 99.70% (335) | thioredoxin reductase TRXB2 |
| M. gilvum PYR-GCK | Mflv_0839 | - | 1e-141 | 76.13% (331) | thioredoxin reductase |
| M. leprae Br4923 | MLBr_02703 | trx | 1e-156 | 83.23% (328) | bifunctional thioredoxin reductase/thioredoxin |
| M. abscessus ATCC 19977 | MAB_4940 | - | 1e-133 | 76.47% (306) | thioredoxin reductase (TrxB) |
| M. marinum M | MMAR_5477 | trxB2 | 1e-164 | 89.44% (322) | thioredoxin reductase TrxB2 |
| M. avium 104 | MAV_5301 | trxB | 1e-151 | 80.70% (342) | thioredoxin-disulfide reductase |
| M. smegmatis MC2 155 | MSMEG_6933 | trxB | 1e-137 | 78.95% (304) | thioredoxin-disulfide reductase |
| M. thermoresistible (build 8) | TH_2413 | trxB2 | 1e-140 | 76.40% (322) | PROBABLE THIOREDOXIN REDUCTASE TRXB2 (TRXR) (TR) |
| M. ulcerans Agy99 | MUL_5066 | trxB2 | 1e-164 | 89.44% (322) | thioredoxin reductase TrxB2 |
| M. vanbaalenii PYR-1 | Mvan_6067 | - | 1e-142 | 77.91% (326) | thioredoxin reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0839|M.gilvum_PYR-GCK -----------------------------MTSSDTVHDVIIIGSGPAGYT
Mvan_6067|M.vanbaalenii_PYR-1 -----------------------------MTSSETIHDVIVIGSGPAGYT
MSMEG_6933|M.smegmatis_MC2_155 -----------------------------MSTSQTVHDVIIIGSGPAGYT
TH_2413|M.thermoresistible__bu ------------------------------MSDQTIHDVIIIGSGPAGYT
MAB_4940|M.abscessus_ATCC_1997 -----------------------MSQAPSTGTESDVHELIIIGSGPAGYT
MAV_5301|M.avium_104 MPAVLFATAGNSHPYPGVYTDPETIARKVFMTADTVHDVIIIGSGPAGYT
Rv3913|M.tuberculosis_H37Rv ----------------------MTAPPVHDRAHHPVRDVIVIGSGPAGYT
Mb3944|M.bovis_AF2122/97 ----------------------MTAPPVHDRAHHPVRDVIVIGSGPAGYT
MMAR_5477|M.marinum_M ----------------------MTT---DSSADATIHDVIVIGSGPAGYT
MUL_5066|M.ulcerans_Agy99 ----------------------MTT---DSSADATIHDVIVIGSGPAGYT
MLBr_02703|M.leprae_Br4923 ----------------------MNT---TPSAHETIHEVIVIGSGPAGYT
: ::::*:*********
Mflv_0839|M.gilvum_PYR-GCK AAVYAARAQLTPLVFEGSQFGGALMTTTEVENYPGFRNGITGPELMDEMR
Mvan_6067|M.vanbaalenii_PYR-1 AAIYAARAQLNPLVFEGSQFGGALMTTTEVENYPGFRNGITGPELMDEMR
MSMEG_6933|M.smegmatis_MC2_155 AAIYAARAQLKPLVFEGTQFGGALMTTTEVENYPGFREGITGPELMDQMR
TH_2413|M.thermoresistible__bu AAIYTARAQLKPLVFEGTQFGGALMTTTEVENYPGFRDGIMGPELMDQMR
MAB_4940|M.abscessus_ATCC_1997 AAVYAARAQLKPIVFEGTQFGGALMTTTEVENYPGFREGIMGPDLMDQMR
MAV_5301|M.avium_104 AALYTARAQLAPVVFEGTSFGGALMTTTEVENYPGFRDGITGPELMDQMR
Rv3913|M.tuberculosis_H37Rv AALYAARAQLAPLVFEGTSFGGALMTTTDVENYPGFRNGITGPELMDEMR
Mb3944|M.bovis_AF2122/97 AALYAARAQLAPLVFEGTSFGGALMTTTGVENYPGFRNGITGPELMDEMR
MMAR_5477|M.marinum_M AALYTARAQLAPLVFEGTSFGGALMTTTEVENYPGFREGITGPELMDEMR
MUL_5066|M.ulcerans_Agy99 AALYTARAQLAPLVFEGTSFGGALMTTTEVENYPGFREGITGPELMDEMR
MLBr_02703|M.leprae_Br4923 AALYAARAQLTPLVFEGTSFGGALMTTTEVENYPGFRNGITGPELMDDMR
**:*:***** *:****:.********* ********:** **:***:**
Mflv_0839|M.gilvum_PYR-GCK EQALRFGADLRMEDVDAVDLTGPVKTVTVGD-ETHSARAVILAMGAAARH
Mvan_6067|M.vanbaalenii_PYR-1 EQALRFGADLRMEDVDAVDLTGPVKSVTVGD-ETFRARAVILAMGAAARH
MSMEG_6933|M.smegmatis_MC2_155 EQALRFGADLRMEDVDAVQLEGPVKTVVVGD-ETHQARAVILAMGAAARH
TH_2413|M.thermoresistible__bu EQALRFGADLRMEDVEAVSLQGPVKTVTVGD-ETHRARAVILAMGAAARH
MAB_4940|M.abscessus_ATCC_1997 EQAIRFGADLRTEDVDEVSLRGPVKTVTVGD-EVYRARAVILAMGAAPRY
MAV_5301|M.avium_104 EQALRFGADLRMEDVESVSLAGPVKSVTTAEGETVRARAVILAMGAAARY
Rv3913|M.tuberculosis_H37Rv EQALRFGADLRMEDVESVSLHGPLKSVVTADGQTHRARAVILAMGAAARY
Mb3944|M.bovis_AF2122/97 EQALRFGADLRMEDVESVSLHGPLKSVVTADGQTHRARAVILAMGAAARY
MMAR_5477|M.marinum_M EQALRFGADLRMEDVESVSLDGPIKSVVTSEGETHRARAVILAMGAAARY
MUL_5066|M.ulcerans_Agy99 EQALRFGADLRMEDVQSVSLDGPIKSVVTSDGETHRARAVILAMGAAARY
MLBr_02703|M.leprae_Br4923 EQALRFGAELRTEDVESVSLRGPIKSVVTAEGQTYQARAVILAMGTSVRY
***:****:** ***: *.* **:*:*...: :. *********:: *:
Mflv_0839|M.gilvum_PYR-GCK LGVPGEDEMLGMGVSTCATCDGFFFRDQDIVVVGGGDSAMEEATFLTRFA
Mvan_6067|M.vanbaalenii_PYR-1 LGVPGEDTLLGMGVSTCATCDGFFFRDQDIAVVGGGDSAMEEATFLTRFA
MSMEG_6933|M.smegmatis_MC2_155 LGVPGEEALTGMGVSTCATCDGFFFRDQDIVVVGGGDSAMEEATFLTRFA
TH_2413|M.thermoresistible__bu LGVPGEEELIGMGVSTCATCDGFFFRDQDIIVVGGGDSAMEEALFLTKFA
MAB_4940|M.abscessus_ATCC_1997 LGVPGEDTLLGRGVSSCATCDGFFFKDQDIAVIGGGDSAMEEATFLTRFA
MAV_5301|M.avium_104 LGVPGEQDLLGRGVSSCATCDGFFFKDQDIAVIGGGDSAMEEATFLTRFA
Rv3913|M.tuberculosis_H37Rv LQVPGEQELLGRGVSSCATCDGFFFRDQDIAVIGGGDSAMEEATFLTRFA
Mb3944|M.bovis_AF2122/97 LQVPGEQELLGRGVSSCATCDGFFFRDQDIAVIGGGDSAMEEATFLTRFA
MMAR_5477|M.marinum_M LHVPGEQELLGRGVSSCATCDGFFFRDQDIAVIGGGDSAMEEATFLTRFA
MUL_5066|M.ulcerans_Agy99 LHVPGEQELLGRGVSSCATCDGFFFRDQDIAVIGGGDSAMEEATFLTRFA
MLBr_02703|M.leprae_Br4923 LQIPGEQELLGRGVSACATCDGSFFRGQDIAVIGGGDSAMEEALFLTRFA
* :***: : * ***:****** **:.*** *:********** ***:**
Mflv_0839|M.gilvum_PYR-GCK RSVTLIHRRDEFRASKIMLERAQQNEKITFLTNTQVLSIEGDPKVTGIRL
Mvan_6067|M.vanbaalenii_PYR-1 RSVTLIHRRDEFRASKIMLERAQANEKITFLTNTQVTAIEGDPKVTGIRL
MSMEG_6933|M.smegmatis_MC2_155 RSVTLIHRRDEFRASKIMLERARANEKITFLTNTEITQIEGDPKVTGVRL
TH_2413|M.thermoresistible__bu RKVTLIHRRDEFRASRIMLERARANEKITFVTNTQVTAIEGDPKVTGVRL
MAB_4940|M.abscessus_ATCC_1997 RSVTLIHRRDEFRASKIMLERAYADPKITVLTNTKIVGVEGTDSVTGLKL
MAV_5301|M.avium_104 RSVTLLHRREEFRASRIMLERARANDKITIVTNKAVEAVEGSETVTGLRL
Rv3913|M.tuberculosis_H37Rv RSVTLVHRRDEFRASKIMLDRARNNDKIRFLTNHTVVAVDGDTTVTGLRV
Mb3944|M.bovis_AF2122/97 RSVTLVHRRDEFRASKIMLDRARNNDKIRFLTNHTVVAVDGDTTVTGLRV
MMAR_5477|M.marinum_M RSVTLVHRRDEFRASKIMINRAQANDKIRILTNKIVLAVDGETTVTGLQL
MUL_5066|M.ulcerans_Agy99 RSVTLVHRRDEFRASKIMTNRAQANDKIRILTNKIVLAVDGETTVTGLQL
MLBr_02703|M.leprae_Br4923 RSVTLVHRRDEFRASKIMLGRARNNDKIKFITNHTVVAVNGYTTVTGLRL
*.***:***:*****:** ** : ** .:** : ::* .***:::
Mflv_0839|M.gilvum_PYR-GCK RNSETGEESRLEVTGVFVAIGHDPRSELVRGQVDVDDEGYVSVQGRTTYT
Mvan_6067|M.vanbaalenii_PYR-1 RNTATGEESTLPVTGVFVAVGHDPRSELVRDQVEVDDAGYVKVQGRTTYT
MSMEG_6933|M.smegmatis_MC2_155 RDTVTGEESKLDVTGVFVAIGHDPRSELVRGQVELDDEGYVKVQGRTTYT
TH_2413|M.thermoresistible__bu RNTVTGEESHMAVTGVFVAIGHDPRSELVRGQLDLDDDGYVLVKGRTTAT
MAB_4940|M.abscessus_ATCC_1997 ENTVTGQASELPVTGMFVAVGHDPRSELVKDVVDVDPDGYVLVRDRSTYT
MAV_5301|M.avium_104 RDTVTGETSTLAVTGVFVAIGHDPRSELVRDVLDTDPDGYVLVQGRTTAT
Rv3913|M.tuberculosis_H37Rv RDTNTGAETTLPVTGVFVAIGHEPRSGLVREAIDVDPDGYVLVQGRTTST
Mb3944|M.bovis_AF2122/97 RDTNTGAETTLPVTGVFVAIGHEPRSGLVREAIDVDPDGYVLVQGRTTST
MMAR_5477|M.marinum_M RDTVTGEETTLPVTGVFVAIGHEPRSSLVRDAVDVDPDGYVLVNGRTTGT
MUL_5066|M.ulcerans_Agy99 RDTVTGEETTLPVTGVFVAIGHEPRSSLVRDAVDVDPDGYVLVNGRTTGT
MLBr_02703|M.leprae_Br4923 RNTTTGEETTLVVTGVFVAIGHEPRSSLVSDVVDIDPDGYVLVKGRTTST
.:: ** : : ***:***:**:*** ** :: * *** *..*:* *
Mflv_0839|M.gilvum_PYR-GCK SVEGVFAAGDLVDHTYRQAITAAGSGCAAAIDAERWLAEHAEP------G
Mvan_6067|M.vanbaalenii_PYR-1 SVEGVFAAGDLVDHTYRQAITAAGSGCAASIDAERWLAEHTEP------G
MSMEG_6933|M.smegmatis_MC2_155 SLDGVFAAGDLVDHTYRQAITAAGSGCAASIDAERWLAEQD---------
TH_2413|M.thermoresistible__bu SVEGVFAAGDLVDRTYRQAITAAGSGCAAAIDAERWLAEQP---------
MAB_4940|M.abscessus_ATCC_1997 SVEGVFAAGDLVDRTYRQAVTAAGSGCAAAIDAERWLAETA---------
MAV_5301|M.avium_104 SIPGVFAAGDLVDRTYRQAVTAAGSGCAAAIDAERWLAEHAES------S
Rv3913|M.tuberculosis_H37Rv SLPGVFAAGDLVDRTYRQAVTAAGSGCAAAIDAERWLAEHA---------
Mb3944|M.bovis_AF2122/97 SLPGVFAAGDLVDRTYRQAVTAAGSGCAAAIDAERWLAEHA---------
MMAR_5477|M.marinum_M SLEGVFAAGDLVDRTYRQAVTAAGSGCAAAIDAERWLAEHE---------
MUL_5066|M.ulcerans_Agy99 SLEGVFAAGDLVDRTYRQAVTAAGSGCAAAIDAERWLAEHE---------
MLBr_02703|M.leprae_Br4923 SMDGVFAAGDLVDRTYRQAITAAGSGCAAAIDAERWLAEHAGSKANETTE
*: **********:*****:*********:*********
Mflv_0839|M.gilvum_PYR-GCK ERTSTTTDDTATDDTDLIGAQQ----------------------------
Mvan_6067|M.vanbaalenii_PYR-1 ERSSTTTDD-----SDLIGAQQ----------------------------
MSMEG_6933|M.smegmatis_MC2_155 --------------------------------------------------
TH_2413|M.thermoresistible__bu -RTPSTEDT------DLIGAPR----------------------------
MAB_4940|M.abscessus_ATCC_1997 -HSQT-----------LIEA------------------------------
MAV_5301|M.avium_104 AAAQGDATEFPGSTDTLIGAPQ----------------------------
Rv3913|M.tuberculosis_H37Rv ATGEADSTD------ALIGAQR----------------------------
Mb3944|M.bovis_AF2122/97 ATGEADSTD------ALIGAQR----------------------------
MMAR_5477|M.marinum_M ATGDADSTD------TLIGAQQ----------------------------
MUL_5066|M.ulcerans_Agy99 ATGDADSTD------TLIGAQQ----------------------------
MLBr_02703|M.leprae_Br4923 ETGDVDSTDTTDWSTAMTDAKNAGVTIEVTDASFFADVLSSNKPVLVDFW
Mflv_0839|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6067|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6933|M.smegmatis_MC2_155 --------------------------------------------------
TH_2413|M.thermoresistible__bu --------------------------------------------------
MAB_4940|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_5301|M.avium_104 --------------------------------------------------
Rv3913|M.tuberculosis_H37Rv --------------------------------------------------
Mb3944|M.bovis_AF2122/97 --------------------------------------------------
MMAR_5477|M.marinum_M --------------------------------------------------
MUL_5066|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02703|M.leprae_Br4923 ATWCGPCKMVAPVLEEIASEQRNQLTVAKLDVDTNPEMAREFQVVSIPTM
Mflv_0839|M.gilvum_PYR-GCK ---------------------------------
Mvan_6067|M.vanbaalenii_PYR-1 ---------------------------------
MSMEG_6933|M.smegmatis_MC2_155 ---------------------------------
TH_2413|M.thermoresistible__bu ---------------------------------
MAB_4940|M.abscessus_ATCC_1997 ---------------------------------
MAV_5301|M.avium_104 ---------------------------------
Rv3913|M.tuberculosis_H37Rv ---------------------------------
Mb3944|M.bovis_AF2122/97 ---------------------------------
MMAR_5477|M.marinum_M ---------------------------------
MUL_5066|M.ulcerans_Agy99 ---------------------------------
MLBr_02703|M.leprae_Br4923 ILFQGGQPVKRIVGAKGKAALLRDLSDVVPNLN