For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HRIFLITVALALLTASPASAITPPPIDPGALPPDVTGPDQPTEQRVLCASPTTLPGSGFHDPPWSNTYLG VADAHKFATGAGVTVAVIDTGVDASPRVPAEPGGDFVDQAGNGLSDCDAHGTLTASIIAGRPAPTDGFVG VAPDARLLSLRQTSEAFEPVGSQANPNDPNATPAAGSIRSLARAVVHAANLGVGVINISEAACYKVSRPI DETSLGASIDYAVNVKGVVVVVAAGNTGGDCVQNPAPDPSTPGDPRGWNNVQTVVTPAWYAPLVLSVGGI GQTGMPSSFSMHGPWVDVAAPAENIVALGDTGEPVNALQGREGPVPIAGTSFAAAYVSGLAALLRQRFPD LTPAQIIHRITATARHPGGGVDDLVGAGVIDAVAALTWDIPPGPASAPYNVRRLPPPVVEPGPDRRPITA VALVAVGLTLALGLGALARRALSRR*
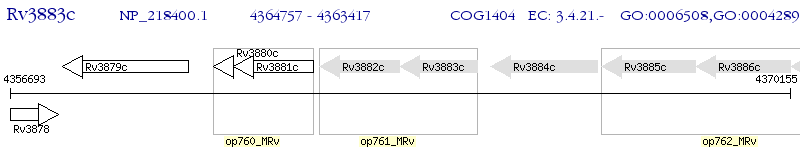
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3883c | mycP1 | - | 100% (446) | MEMBRANE-ANCHORED MYCOSIN MYCP1 (SERINE PROTEASE) (SUBTILISIN-LIKE PROTEASE) (SUBTILASE-LIKE) (MYCOSIN-1) |
| M. tuberculosis H37Rv | Rv0291 | mycP3 | 4e-92 | 43.56% (450) | membrane-anchored mycosin |
| M. tuberculosis H37Rv | Rv3449 | mycP4 | 2e-85 | 44.89% (450) | membrane-anchored mycosin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3913c | - | 0.0 | 99.78% (446) | secreted protease |
| M. gilvum PYR-GCK | Mflv_0755 | - | 0.0 | 68.93% (441) | peptidase S8 and S53, subtilisin, kexin, sedolisin |
| M. leprae Br4923 | MLBr_00041 | - | 0.0 | 79.37% (446) | putative secreted protease |
| M. abscessus ATCC 19977 | MAB_3758 | - | 3e-96 | 46.91% (437) | putative protease |
| M. marinum M | MMAR_5459 | mycP3_1 | 0.0 | 86.10% (446) | membrane-anchored serine protease (mycosin), MycP3_1 |
| M. avium 104 | MAV_4862 | - | 4e-97 | 44.09% (465) | subtilase family protein |
| M. smegmatis MC2 155 | MSMEG_0083 | - | 0.0 | 72.46% (443) | membrane-anchored mycosin mycp1 |
| M. thermoresistible (build 8) | TH_3348 | - | 0.0 | 71.11% (443) | PUTATIVE peptidase S8 and S53, subtilisin, kexin, sedolisin |
| M. ulcerans Agy99 | MUL_1213 | mycP3 | 2e-94 | 43.98% (457) | membrane-anchored mycosin MycP3 |
| M. vanbaalenii PYR-1 | Mvan_0092 | - | 0.0 | 71.21% (448) | peptidase S8 and S53, subtilisin, kexin, sedolisin |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3883c|M.tuberculosis_H37Rv ----------------------MHRIFLITVALALLT-----ASPASAIT
Mb3913c|M.bovis_AF2122/97 ----------------------MHRIFLITVALALLT-----ASPASAIT
MMAR_5459|M.marinum_M MQAGLTRACQSFTAARERSDSGVHRTLLTMVALALLT-----APPALAID
MLBr_00041|M.leprae_Br4923 ----------------------MHRILLMMVALALLY-----APPALAIN
Mflv_0755|M.gilvum_PYR-GCK --------------------MRRIGALIAALLLVLVT-----APPAGAVE
Mvan_0092|M.vanbaalenii_PYR-1 --------------------MRRIGALIAALFLVLLT-----APPASAIE
TH_3348|M.thermoresistible__bu -----------------------LGVLITALLLAVVT-----APPAGAIE
MSMEG_0083|M.smegmatis_MC2_155 --------------------MQRVAVMVLAVLLALFS-----APPAWAID
MAB_3758|M.abscessus_ATCC_1997 -----------MSLGVARRIATAGAVVMLAMSSQLATGSMHVMATAHAFA
MAV_4862|M.avium_104 -------------------MIRPASACLAAVLALLPATATWASPPALAIS
MUL_1213|M.ulcerans_Agy99 -------------------MKRAGSACLTAILAIFTTGALWAAPPVSAIS
: : . ... *.
Rv3883c|M.tuberculosis_H37Rv PPPIDPGALP-PDVTGPDQPTEQRVLCASPTTLPGSGFHDPPWSNTYLGV
Mb3913c|M.bovis_AF2122/97 PPPIDPGALP-PDVAGPDQPTEQRVLCASPTTLPGSGFHDPPWSNTYLGV
MMAR_5459|M.marinum_M PPSIDPGAVP-PDVTGPDQPTEQRVLCTSPTTLPDSSFHDPPWSNAYMGV
MLBr_00041|M.leprae_Br4923 PPSIDPGAVP-PDVTGPDQPTEQKVMCSAPTTLPDSSFHDPPWSNIYIGV
Mflv_0755|M.gilvum_PYR-GCK QPVIDPAAIP-PDETGPDQPMEQRRVCAAPTVFPNANFADRPWASDYLRL
Mvan_0092|M.vanbaalenii_PYR-1 PPIVDPAAIP-PDETGPDQPMEQRRICAAPTVFPNSNFADRPWASDYLRL
TH_3348|M.thermoresistible__bu PPVIDPAAVP-PDETGPDNPMEQRRVCAAPTVYPDSNFADRPWASDYLRL
MSMEG_0083|M.smegmatis_MC2_155 PPVIDAGAVP-PDETGPDQPTEQRKICATPTVMPNSNFADRPWANDYLRI
MAB_3758|M.abscessus_ATCC_1997 PLPVDPSMLPGPRPPAPIEPTEQKELCRFPGIDPKRDLNKVPTQQRGLDL
MAV_4862|M.avium_104 PPTVDPGVPPPSGAPGPGQPMEQRAPCTTSGVIPGSEPAAVTPSQSALNL
MUL_1213|M.ulcerans_Agy99 PPAVDDSVQPPSGTPGPVQTMEQRGACSVSGLIPGTDISVPGASQAMLNL
:* . * . ..* :. **: * . * . : :
Rv3883c|M.tuberculosis_H37Rv ADAHKFATGAGVTVAVIDTGVDASPRVP-AEPGGDFVDQAGNGLSDCDAH
Mb3913c|M.bovis_AF2122/97 ADAHKFATGAGVTVAVIDTGVDASPRVP-AEPGGDFVDQAGNGLSDCDAH
MMAR_5459|M.marinum_M GEAHKFATGAGVTVAVIDTGVDASPRVP-AEPGGDFVDQAGDGLSDCDAH
MLBr_00041|M.leprae_Br4923 SEAHKFATGAGVTVAVIDTGVEASPRVP-AEPGGDFVDQAGNGLSDCDAH
Mflv_0755|M.gilvum_PYR-GCK ADAQQFATGAGVTVAVIDTGVNGSPRVP-AEPGGDFVDAAGDGMSDCDAH
Mvan_0092|M.vanbaalenii_PYR-1 TEAQKFATGAGVTVAVIDTGVNGSPRVP-AEPGGDFVDQAGNGMSDCDAH
TH_3348|M.thermoresistible__bu TEAHKFATGAGITVAVIDTGVNGSPRVP-AEPGGDFVDAAGNGMSDCDAH
MSMEG_0083|M.smegmatis_MC2_155 QEAQKFATGAGVTVAVIDTGVNGSPRVP-AEPGGDFVDAAGNGMSDCDAH
MAB_3758|M.abscessus_ATCC_1997 PLVWQLSRGAGQTVAVIDTGVARSPRLPPLIPGGDYVFEGGNGTQDCDAH
MAV_4862|M.avium_104 PAAWRFSRGDGQLVAVLDTGVRPGPRLPNVEPGGDFVETT-DGLTDCDGH
MUL_1213|M.ulcerans_Agy99 PAAWQFSRGEGQLVAILDTGVQPGPRLPNVDAGGDFVDST-DGLTDCDGR
. ::: * * **::**** .**:* .***:* :* ***.:
Rv3883c|M.tuberculosis_H37Rv GTLTASIIAGRPAPT--DGFVGVAPDARLLSLRQTSEAFEPVG-SQANPN
Mb3913c|M.bovis_AF2122/97 GTLTASIIAGRPAPT--DGFVGVAPDARLLSLRQTSEAFEPVG-SQANPN
MMAR_5459|M.marinum_M GTLTASIIGGRPAPT--DGFVGVAPDVRLLSLRQTSEAFEPVG-SQPNPN
MLBr_00041|M.leprae_Br4923 GTLTASIIGGRPAPT--DGFIGVAPDVRLLSLRQTSEAFQPVG-VQPNAN
Mflv_0755|M.gilvum_PYR-GCK GTLTAAVIAGRPAPT--DAFIGVAPDARILSLRQTSDSFSPVG-ARNDPN
Mvan_0092|M.vanbaalenii_PYR-1 GTLTAAVIAGRPAPH--DAFIGVAPNARILSLRQTSDSFLPVG-ARTDPN
TH_3348|M.thermoresistible__bu GTLTASVIAGRGAPT--DGFIGVAPDARILSLRHTSAAFQPVG-ARTDPN
MSMEG_0083|M.smegmatis_MC2_155 GTMTAAIIGGRPSPT--DGFVGMAPDVRLLSLRQTSVAFQPKG-ARQDPN
MAB_3758|M.abscessus_ATCC_1997 GTIVAGIIAAAPDPTGASAFSGIAPDVSLITIRQSSAKFAVKNKGGNDRD
MAV_4862|M.avium_104 GTLLAGLVAGQPSPD--DGFAGVAPAARVVSIRTTSAKFSPRT----PGG
MUL_1213|M.ulcerans_Agy99 GTLVAGIVAGQPG-N--DGFSGVAPAARLLSIRVTSDKFSARS----SGG
**: *.::.. ..* *:** . ::::* :* * .
Rv3883c|M.tuberculosis_H37Rv DPNATPAAGSIRSLARAVVHAANLGVGVINISEAACYKVSRPIDETSLGA
Mb3913c|M.bovis_AF2122/97 DPNATPAAGSIRSLARAVVHAANLGVGVINISEAACYKVSRPIDETSLGA
MMAR_5459|M.marinum_M DPNATPAAGSIRSLARAVVHAANLGAGVINISEAACYKVSRPIDEISLGA
MLBr_00041|M.leprae_Br4923 DPNAIPAAGSIRSLARAVVHAANLGASVINISETACYKANKPLDESTLGA
Mflv_0755|M.gilvum_PYR-GCK DPNATQTAGSLRSLARAIVHAANLGAQVINISEAACYKVTRRIDEAVVGA
Mvan_0092|M.vanbaalenii_PYR-1 DPNSTQTAGSLRSLARAIVHAANMGAQVINISEAACYKITRPINEAVVGA
TH_3348|M.thermoresistible__bu NPNTTQTAGSLRSLARAIVHAANLGAQVINISEAACYKVTRPIDETGVGA
MSMEG_0083|M.smegmatis_MC2_155 DPNTTQTAGSIRSLARSVVHAANLGAQVINISEAACYKVTRRIDETSLGA
MAB_3758|M.abscessus_ATCC_1997 DPSKISGVGNVDTLAMAVRTAADMGATVINISEVACASAKQFLDDRPLGA
MAV_4862|M.avium_104 DPLLARAAAEVGTLARAIVHAADLGARVIDIGSATCLPVDRPVDQAALGA
MUL_1213|M.ulcerans_Agy99 DPSVERATIDVAALGRAIVHAADRGARVINISSITCLPADRQVDQAALGA
:* . .: :*. :: **: *. **:*.. :* : ::: :**
Rv3883c|M.tuberculosis_H37Rv SIDYAVNVKGVVVVVAAGNTG-------GDCV-QNPAPDPSTPGDPRGWN
Mb3913c|M.bovis_AF2122/97 SIDYAVNVKGVVVVVAAGNTG-------GDCV-QNPAPDPSTPGDPRGWN
MMAR_5459|M.marinum_M AIDYAVNAKNAVVVVAAGNTG-------GDCS-QNPMPDASTPNDPRGWN
MLBr_00041|M.leprae_Br4923 AINYAVNVKGVVIIVAAGNTG-------GDCT-QNPMPDVSMPNDPRGWS
Mflv_0755|M.gilvum_PYR-GCK AINYAVNVKGTVIVAAAGNTG-------ENCA-QNPPPDPAVPADPRGWQ
Mvan_0092|M.vanbaalenii_PYR-1 AINYAVNVKGAVIVVAAGNTG-------ENCT-QNPPPDPAVPADPRGWQ
TH_3348|M.thermoresistible__bu AVNYAVHVKNAVVIAAAGNTG-------QDCT-QNPPPDPAVPSDPRGWQ
MSMEG_0083|M.smegmatis_MC2_155 AINYAVNVKGAVIVVAAGNTG-------QDCS-QNPPPDPSVPSDPRGWR
MAB_3758|M.abscessus_ATCC_1997 ALQYAVDVKNVVVVVAAGNVGD------GQCKSQNPPPDPARPNTP-DWD
MAV_4862|M.avium_104 AVRYAAVDKDAVIVAAAGDSGPTGSFAGGTSCESNPLTDLSRPSDPRNWA
MUL_1213|M.ulcerans_Agy99 AIRYAAVDKDVVIVAAAGNSGAGGSGSGGT-CESNPLTDLSHPDDPRNWA
:: **. *..*::.***: * .** .* : * * .*
Rv3883c|M.tuberculosis_H37Rv NVQTVVTPAWYAPLVLSVGGIGQTGMPSSFSMHGPWVDVAAPAENIVALG
Mb3913c|M.bovis_AF2122/97 NVQTVVTPAWYAPLVLSVGGIGQTGMPSSFSMHGPWVDVAAPAENIVALG
MMAR_5459|M.marinum_M KVQTVVTPAWYAPLVLTVGGIGQNGVPSSFSMHGPWVGVAAPAENIIALG
MLBr_00041|M.leprae_Br4923 KVQTIATPAWYAPLVLTVGGIAQNGMPSQFSVHGPWVSVAAPTENITTLG
Mflv_0755|M.gilvum_PYR-GCK QVQTIVSPAWYSPLVLTVGSIAPNGQPSGFSVQGPWVGAAAPGENLVALG
Mvan_0092|M.vanbaalenii_PYR-1 EVQTIVSPAWYSPLVLTVGSIAPNGQPSGFSMQGPWVGAAAPGENLIALG
TH_3348|M.thermoresistible__bu QVQTIVSPAWYSPLVLTVGGIGPTGQPSNFSMSGPWVGAAAPAENITALG
MSMEG_0083|M.smegmatis_MC2_155 EVQTIVSPAWYDPLVLTVGSIGQNGQPSNFSMSGPWVGAAAPGENLTSLG
MAB_3758|M.abscessus_ATCC_1997 SVLETVSPAWYNDLVLTVGSIGPDGQPSKFSLGGPWVDVSAPGEDIVSLD
MAV_4862|M.avium_104 GVTSLSIPSWWQPYVLSVASLTPDGQPSKFTVAGPWVGVAAPGERIVSIG
MUL_1213|M.ulcerans_Agy99 GVTSVSIPSLWQPYVLSVASLTAGGQPSKFTMAGPWVGIAAPGENIVSVS
* *: : **:*..: * ** *:: ****. :** * : ::.
Rv3883c|M.tuberculosis_H37Rv --DTGEPVNALQGR-EGPVPIAGTSFAAAYVSGLAALLRQRFPDLTPAQI
Mb3913c|M.bovis_AF2122/97 --DTGEPVNALQGR-EGPVPIAGTSFAAAYVSGLAALLRQRFPDLTPAQI
MMAR_5459|M.marinum_M --DHGEPVNALQGR-EGPVPIAGTSFAAAYVSGLAALVRQRFPELTPVQV
MLBr_00041|M.leprae_Br4923 --YNGEPVNALPGE-EGPVALAGTSFATAYVSGLAALLRQRFPDLTPAQI
Mflv_0755|M.gilvum_PYR-GCK --YDGNPVNALQGE-DGPIPISGTSFSAAFVSGVAALIKQRFPELTPGQI
Mvan_0092|M.vanbaalenii_PYR-1 --YDGNPVNALQGE-DGPIPISGTSFSAAYVSGLAALLRQRFPELTPAQI
TH_3348|M.thermoresistible__bu --YGGEPVNALQGQ-DGLVPVAGTSFAAAYVSGLAALIRQRYPDLTPAQV
MSMEG_0083|M.smegmatis_MC2_155 --YDGQPVNATPGE-DGPVPLNGTSFSAAYVSGLAALVKQRFPDLTPAQI
MAB_3758|M.abscessus_ATCC_1997 P-NRDALTNAYMGD-QGPVPIQGTSFSAPVVSGVVALVRSRFPQLSAREV
MAV_4862|M.avium_104 NGDGAGLTNGLPDEHQQLAALNGTGYAAGYVAGVAALVRSKYPDLSATAV
MUL_1213|M.ulcerans_Agy99 NADGGGLANGLPNQRQQLTPLSGTSYAAGYVSGVATLVRSNYPELTAAQV
.*. . : .: **.::: *:*:.:*::..:*:*:. :
Rv3883c|M.tuberculosis_H37Rv IHRITATARHPGGGVDDLVGAGVIDAVAALTWDIPPGPASAPYN---VRR
Mb3913c|M.bovis_AF2122/97 IHRITATARHPGGGVDDLVGAGVIDAVAALTWDIPPGPASAPYN---VRR
MMAR_5459|M.marinum_M MNRITATARHPGGGIDNLVGAGVVNAVAALTWDIPPGPASVPPS---VRR
MLBr_00041|M.leprae_Br4923 INRIIATARHPGGGVDDIVGAGVIDAVAALTWDITPGSASELFN---VRS
Mflv_0755|M.gilvum_PYR-GCK INRITATARHPGGGVNNVVGAGVIDPVAALTWDVPAGPADIPYQ---IKE
Mvan_0092|M.vanbaalenii_PYR-1 INRITATARHPGGGVNNVVGAGVIDPVAALTWDVPPGPADIPYN---IKE
TH_3348|M.thermoresistible__bu INRITATARHPGGGVDNYVGAGVIDPVAALTWDVPEGPETAPYR---AKE
MSMEG_0083|M.smegmatis_MC2_155 INRITATARHPGGGVDNYVGAGVIDPVAALTWEIPDGPEKAPFR---VKE
MAB_3758|M.abscessus_ATCC_1997 MDRIKNTAHRPPGGWDPYLGFGVVDALAAVSDDLGTTSDLARAN--AAKK
MAV_4862|M.avium_104 VHRITATAHNGARDPSNVVGAGSVDPVAALTWQLPAGDQPGP---A-AKP
MUL_1213|M.ulcerans_Agy99 MHRLTATAHNGARDPSNVVGAGILDPAAALTWELPAADGGAPQVAP-AKP
:.*: **:. . . :* * ::. **:: :: :
Rv3883c|M.tuberculosis_H37Rv LPPPVVEPGPDRRP----ITAVALVAVGLTLALGLGALARRALSRR----
Mb3913c|M.bovis_AF2122/97 LPPPVVEPGPDRRP----ITAVALVAVGLTLALGLGALARRALSRR----
MMAR_5459|M.marinum_M LPPPRIEPGPDHRP----ITMVAVSVLGLTLVLGLGTLAARALRRR----
MLBr_00041|M.leprae_Br4923 VPPPVITPGPDRNT----IMIVAQGILGLSIALGLGVLVRRALQRQ----
Mflv_0755|M.gilvum_PYR-GCK IPPPVHIPPPDRSP----ITGVVVVTIVVAALLGIGAMARRALRRR----
Mvan_0092|M.vanbaalenii_PYR-1 IPPPVYVPPPDRSP----ITGVVLVGVGLAALLGIAALGRRALRRR----
TH_3348|M.thermoresistible__bu IPEPEFIPPPDRGP----ITWVVVSSAAVALALGIGALTRRALRRR----
MSMEG_0083|M.smegmatis_MC2_155 VPPPVYIPPPDRGP----ITAVVIAGATLAFALGIGALARRALRRKQ---
MAB_3758|M.abscessus_ATCC_1997 LVLPPAPPPPDPRPRRVALIGIAVCAAFLAAALAMLAPVRRLRQSREHPD
MAV_4862|M.avium_104 IAVPPAPAPPDTTPRTVAVAGTAALAVLVAVAGAVTALAARRR--KDPTA
MUL_1213|M.ulcerans_Agy99 VSVPPAPQPRNTTPRNVAFAGAAALTVLVAGVAATVAIARRRKDAEGRTA
: * : . . . :: . . * .
Rv3883c|M.tuberculosis_H37Rv ---
Mb3913c|M.bovis_AF2122/97 ---
MMAR_5459|M.marinum_M ---
MLBr_00041|M.leprae_Br4923 ---
Mflv_0755|M.gilvum_PYR-GCK ---
Mvan_0092|M.vanbaalenii_PYR-1 ---
TH_3348|M.thermoresistible__bu ---
MSMEG_0083|M.smegmatis_MC2_155 ---
MAB_3758|M.abscessus_ATCC_1997 E--
MAV_4862|M.avium_104 ---
MUL_1213|M.ulcerans_Agy99 REL