For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRKNVTRTITAIAVVVLLGWSFFYFSDDTRGYKPVDTSVAITQINGDNVKSAQIDDREQQLRLILKKGNN ETDGSEKVITKYPTGYAVDLFNALSAKNAKVSTVVNQGSILGELLVYVLPLLLLVGLFVMFSRMQGGARM GFGFGKSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIPKGVLLYGPPGTGKTL LARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRDLFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDE REQTLNQLLVEMDGFGDRAGVILIAATNRPDILDPALLRPGRFDRQIPVSNPDLAGRRAVLRVHSKGKPM AADADLDGLAKRTVGMTGADLANVINEAALLTARENGTVITGPALEEAVDRVIGGPRRKGRIISEQEKKI TAYHEGGHTLAAWAMPDIEPIYKVTILARGRTGGHAVAVPEEDKGLRTRSEMIAQLVFAMGGRAAEELVF REPTTGAVSDIEQATKIARSMVTEFGMSSKLGAVKYGSEHGDPFLGRTMGTQPDYSHEVAREIDEEVRKL IEAAHTEAWEILTEYRDVLDTLAGELLEKETLHRPELESIFADVEKRPRLTMFDDFGGRIPSDKPPIKTP GELAIERGEPWPQPVPEPAFKAAIAQATQAAEAARSDAGQTGHGANGSPAGTHRSGDRQYGSTQPDYGAP AGWHAPGWPPRSSHRPSYSGEPAPTYPGQPYPTGQADPGSDESSAEQDDEVSRTKPAHG*
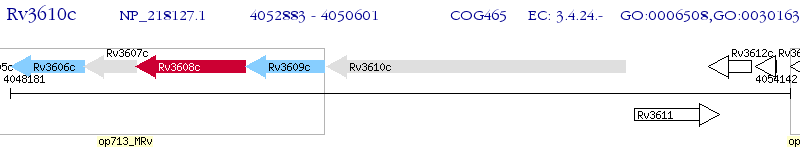
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3610c | ftsH | - | 100% (760) | MEMBRANE-BOUND PROTEASE FTSH (CELL DIVISION PROTEIN) |
| M. tuberculosis H37Rv | Rv0435c | - | 6e-44 | 39.91% (228) | putative ATPase |
| M. tuberculosis H37Rv | Rv2115c | - | 4e-31 | 35.50% (231) | ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3640c | ftsH | 0.0 | 100.00% (760) | membrane-bound protease FTSH (cell division protein) |
| M. gilvum PYR-GCK | Mflv_1416 | - | 0.0 | 80.67% (776) | ATP-dependent metalloprotease FtsH |
| M. leprae Br4923 | MLBr_00222 | ftsH | 0.0 | 87.22% (790) | putative integral membrane peptidase |
| M. abscessus ATCC 19977 | MAB_0533 | - | 0.0 | 81.30% (754) | cell division protein FtsH-like protein |
| M. marinum M | MMAR_5113 | ftsH | 0.0 | 88.48% (764) | membrane-bound protease FtsH |
| M. avium 104 | MAV_0542 | hflB | 0.0 | 85.41% (802) | ATP-dependent metallopeptidase HflB |
| M. smegmatis MC2 155 | MSMEG_6105 | - | 0.0 | 83.44% (761) | cell division protein |
| M. thermoresistible (build 8) | TH_0991 | ftsH | 0.0 | 82.02% (762) | MEMBRANE-BOUND PROTEASE FTSH (CELL DIVISION PROTEIN) |
| M. ulcerans Agy99 | MUL_4192 | ftsH | 0.0 | 88.57% (761) | membrane-bound protease FtsH |
| M. vanbaalenii PYR-1 | Mvan_5372 | - | 0.0 | 83.88% (757) | ATP-dependent metalloprotease FtsH |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3610c|M.tuberculosis_H37Rv ---MNRKNVTRTITAIAVVVLLGWSFFYFSDDTRGYKPVDTSVAITQING
Mb3640c|M.bovis_AF2122/97 ---MNRKNVTRTITAIAVVVLLGWSFFYFSDDTRGYKPVDTSVAITQING
MLBr_00222|M.leprae_Br4923 MLLMNRKNVIRMVTAIAVVVLLGWSFFYFSDDTRGYKFVDTSVAMSQING
MAV_0542|M.avium_104 ---MNRKNVIRTITAIAVVVLLGWSFFYFSDDTRGYKPVDTSVAMAQING
MMAR_5113|M.marinum_M ---MNRKNVIRTVTVVAVVVVLGWSFFYFSDDTRGYKPVDTSVAMSQINA
MUL_4192|M.ulcerans_Agy99 ---MNRKNVIRTVTAVAVVVVLGWSFFYFSDDTRGYKPVDTSVAMSQINA
Mflv_1416|M.gilvum_PYR-GCK ---MNRKNVIRTLTVIAVVLLLGWSFFYFSDDTRGYKPVDTTIAIAQING
Mvan_5372|M.vanbaalenii_PYR-1 ---MNRKNVIRTLTVIAVVLLLGWSFFYFSDDTRGYKPVDTSVAMAQISG
MSMEG_6105|M.smegmatis_MC2_155 ---MNRKNVIRTLTVIGVVLLLGWSFFYFSDDTRGYKPVDTTVALAQIRG
TH_0991|M.thermoresistible__bu ---MNRKTVIRTLTIVAVVLLLGWSFFYFGDDTREYKPVDTSVALAQIQA
MAB_0533|M.abscessus_ATCC_1997 ---MNRTNVFRTLGVIVVVLLLGWSFFYFSDDTRGYKPVDTTVAIKQIDT
***..* * : : **::********.**** ** ***::*: **
Rv3610c|M.tuberculosis_H37Rv DNVKSAQIDDREQQLRLILKKGNNETDGSEKVITKYPTGYAVDLFNALSA
Mb3640c|M.bovis_AF2122/97 DNVKSAQIDDREQQLRLILKKGNNETDGSEKVITKYPTGYAVDLFNALSA
MLBr_00222|M.leprae_Br4923 HNVKSAQIDDREQQLRLTLKKGNNDTDGSDKVITKYPTGYAVDLFNALSA
MAV_0542|M.avium_104 DNVKSAQIDDREQQLRLTLKKGNGDTDNSDKVITKYPSGYAVDLFNALSA
MMAR_5113|M.marinum_M DNVKSAQIDDREQQLRLVLKKSSSDTDGSDKVITKYPTGYAVDLFNALTA
MUL_4192|M.ulcerans_Agy99 DNAKSAQIDDREQQLRLVLKKSSSDTDGSDKVITKYPTGYAVDLFNALTA
Mflv_1416|M.gilvum_PYR-GCK DNVNSAQIDDREQQLRLELKNGNGDTENSTQILTKYPTGYGATLFESLQD
Mvan_5372|M.vanbaalenii_PYR-1 DNVKSAQIDDREQQLRLELKNGNGDTENSDKIITKYPTGYGVPLFEALQA
MSMEG_6105|M.smegmatis_MC2_155 DNVNSAQIDDREQQVRLDLKSGNGDTENSDKIITKYPTGYGVTLFNALQD
TH_0991|M.thermoresistible__bu DNVDSAQIDDREQQVRLDLKNANSDTGDSDKIIAKYPTGYAVPLFDALQD
MAB_0533|M.abscessus_ATCC_1997 DNVKSARIDDREQQLRLDLKAANGDTEGKDKVITKYPTGYGVPLLEKLNG
.*..**:*******:** ** ...:* .. ::::***:**.. *:: *
Rv3610c|M.tuberculosis_H37Rv KNAKVSTVVNQGSILGELLVYVLPLLLLVGLFVMFSRMQGGAR-MGFGFG
Mb3640c|M.bovis_AF2122/97 KNAKVSTVVNQGSILGELLVYVLPLLLLVGLFVMFSRMQGGAR-MGFGFG
MLBr_00222|M.leprae_Br4923 KNTKVTTAVNEGSILGELLVYVLPLLLLVGLFVMFSRMQGGAR-MGFGFG
MAV_0542|M.avium_104 KNAKVSTVVNQGSILGELLVYVLPLLLLVGLFVMFSRMQGGAR-MGFGFG
MMAR_5113|M.marinum_M KNAKVSTVVNQGSILGELLVYVLPLLLLVGLFVMFSRMQGGAR-MGFGFG
MUL_4192|M.ulcerans_Agy99 KNAKVSTVVNQGSILGELLVYVLPLLLLVGLFVMFSRMQGGAR-MGFGFG
Mflv_1416|M.gilvum_PYR-GCK KNIKTNTVVNQSSIFGTLLIYMLPLLLLVALFVLFSRMQSGGR-MGFGFG
Mvan_5372|M.vanbaalenii_PYR-1 KNTKINTVVNQSSILGTLLIYMLPLLLLVGLFVMFSRMQTGGR-MGFGFG
MSMEG_6105|M.smegmatis_MC2_155 KGAKTNTVVNQGSVFGTLLIYMLPLLLLVALFVMFSRMQTGGR-MGFGFG
TH_0991|M.thermoresistible__bu KNAKVNTVVNQGSVLGTLLIYMLPLLLLVALFVFFMRMQSGGR-MGFGFG
MAB_0533|M.abscessus_ATCC_1997 KGAAVNTTVNQGSILGSLLLYLLPVILLVVLFFAFSRMQSGGRGMGFGFG
*. .*.**:.*::* **:*:**::*** **. * *** *.* ******
Rv3610c|M.tuberculosis_H37Rv KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
Mb3640c|M.bovis_AF2122/97 KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
MLBr_00222|M.leprae_Br4923 KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPCRYQTLGAKIP
MAV_0542|M.avium_104 KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPGRYQALGAKIP
MMAR_5113|M.marinum_M KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
MUL_4192|M.ulcerans_Agy99 KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
Mflv_1416|M.gilvum_PYR-GCK KSKAKMLSKDMPRTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
Mvan_5372|M.vanbaalenii_PYR-1 KSKAKMLGKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
MSMEG_6105|M.smegmatis_MC2_155 KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
TH_0991|M.thermoresistible__bu KSKAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
MAB_0533|M.abscessus_ATCC_1997 KSRAKQLSKDMPKTTFADVAGVDEAVEELYEIKDFLQNPSRYQALGAKIP
**:** *.****:************************** ***:******
Rv3610c|M.tuberculosis_H37Rv KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
Mb3640c|M.bovis_AF2122/97 KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
MLBr_00222|M.leprae_Br4923 KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
MAV_0542|M.avium_104 KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
MMAR_5113|M.marinum_M KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
MUL_4192|M.ulcerans_Agy99 KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
Mflv_1416|M.gilvum_PYR-GCK KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
Mvan_5372|M.vanbaalenii_PYR-1 KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
MSMEG_6105|M.smegmatis_MC2_155 KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
TH_0991|M.thermoresistible__bu KGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
MAB_0533|M.abscessus_ATCC_1997 RGVLLYGPPGTGKTLLARAVAGEAGVPFFTISGSDFVEMFVGVGASRVRD
:*************************************************
Rv3610c|M.tuberculosis_H37Rv LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
Mb3640c|M.bovis_AF2122/97 LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
MLBr_00222|M.leprae_Br4923 LFDQAKQNSPCIIFVDEIDAVGRQRGTGLGGGHDEREQTLNQLLVEMDGF
MAV_0542|M.avium_104 LFDQAKQNNPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
MMAR_5113|M.marinum_M LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
MUL_4192|M.ulcerans_Agy99 LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
Mflv_1416|M.gilvum_PYR-GCK LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
Mvan_5372|M.vanbaalenii_PYR-1 LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
MSMEG_6105|M.smegmatis_MC2_155 LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
TH_0991|M.thermoresistible__bu LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
MAB_0533|M.abscessus_ATCC_1997 LFEQAKQNSPCIIFVDEIDAVGRQRGAGLGGGHDEREQTLNQLLVEMDGF
**:*****.*****************:***********************
Rv3610c|M.tuberculosis_H37Rv GDRAGVILIAATNRPDILDPALLRPGRFDRQIPVSNPDLAGRRAVLRVHS
Mb3640c|M.bovis_AF2122/97 GDRAGVILIAATNRPDILDPALLRPGRFDRQIPVSNPDLAGRRAVLRVHS
MLBr_00222|M.leprae_Br4923 GDRAGVILIAATNRPDILDPALLRPGRFDRQIPVSNPDLAGRRAVLRVHS
MAV_0542|M.avium_104 DPRAGVILIAATNRPDILDPALLRPGRFDRQIPVSNPDLAGRRAVLAVHS
MMAR_5113|M.marinum_M GDRAGVILIAATNRPDILDPALLRPGRFDRQIPVTNPDLAGRRAVLRVHS
MUL_4192|M.ulcerans_Agy99 GDRAGVILIAATNRPDILDPALLRPGRFDRQIPVTNPDLAGRRAVLRVHS
Mflv_1416|M.gilvum_PYR-GCK GDRQGVILIAATNRPDILDPALLRPGRFDRQIPVSSPDLAGRRAVLKVHS
Mvan_5372|M.vanbaalenii_PYR-1 GDRQGVILIAATNRPDILDPALLRPGRFDRQIPVSNPDLAGRRAVLRVHS
MSMEG_6105|M.smegmatis_MC2_155 GDRQGVILIAATNRPDILDPALLRPGRFDRQIPVSNPDLAGRRAVLKVHS
TH_0991|M.thermoresistible__bu GDRQGVILIAATNRPDILDPALLRPGRFDRQIPVTNPDLAGRKAVLRVHA
MAB_0533|M.abscessus_ATCC_1997 GDRQGVILIAATNRPDILDPALLRPGRFDRQIPVSSPDLAGRKAVLKVHS
. * ******************************:.******:*** **:
Rv3610c|M.tuberculosis_H37Rv KGKPMAADADLDGLAKRTVGMTGADLANVINEAALLTARENGTVITGPAL
Mb3640c|M.bovis_AF2122/97 KGKPMAADADLDGLAKRTVGMTGADLANVINEAALLTARENGTVITGPAL
MLBr_00222|M.leprae_Br4923 KGKPIADDADLDGLAKRTVGMTGADLANVVNEAALLTARENGLVITGPAL
MAV_0542|M.avium_104 KGKPIAPDADLDGLAKRTVGMTGADLANVINEAALLTARENGTVITSAAL
MMAR_5113|M.marinum_M KGKPIADGADLDGLAKRTVGMTGADLANVINEAALLTARENGTVITGPAL
MUL_4192|M.ulcerans_Agy99 KGKPIADGADLDGLAKRTVGMTGADLANVINEAALLTARENGTVITGPAL
Mflv_1416|M.gilvum_PYR-GCK QGKPIAEDADLEGLAKRTVGMSGADLANVINEAALLTARENGTIITGPAL
Mvan_5372|M.vanbaalenii_PYR-1 QGKPIADDADLDGLAKRTVGMSGADLANVINEAALLTARENGTIITGPAL
MSMEG_6105|M.smegmatis_MC2_155 QGKPIAPDADLDGLAKRTVGMSGADLANVINEAALLTARENGTVITGPAL
TH_0991|M.thermoresistible__bu QGKPLAPDVDLDGLAKRTVGMSGADLANVINEAALLTARENGTMITGPAL
MAB_0533|M.abscessus_ATCC_1997 AGKPFGPDVDFDGLAKRTVGMSGADLANVINEAALLTARENGTVITAAAL
***:. ..*::*********:*******:************ :**..**
Rv3610c|M.tuberculosis_H37Rv EEAVDRVIGGPRRKGRIISEQEKKITAYHEGGHTLAAWAMPDIEPIYKVT
Mb3640c|M.bovis_AF2122/97 EEAVDRVIGGPRRKGRIISEQEKKITAYHEGGHTLAAWAMPDIEPIYKVT
MLBr_00222|M.leprae_Br4923 EEAVDRVIGGPRRKGRIISEQEKKITAYHEGGHTLAAWAMPDIEPIYKVT
MAV_0542|M.avium_104 EEAVDRVIGGPRRKGRIISEQEKKITAYHEGGHTLAAWAMPDIEPIYKVT
MMAR_5113|M.marinum_M EEAVDRVIGGPRRKGRIISEHEKKITAYHEGGHTLAAWAMPDIDPVYKVT
MUL_4192|M.ulcerans_Agy99 EEAVDRVIGGPRRKGRIISEHEKKITAYHEGGHTLAAWAMPDIDPVYKVT
Mflv_1416|M.gilvum_PYR-GCK EEAVDRVVGGPRRKSRIISEHEKKITAYHEGGHTLAAWAMPDIEPIYKVT
Mvan_5372|M.vanbaalenii_PYR-1 EEAVDRVVGGPRRKSRIISEHEKKITAYHEGGHTLAAWAMPDIDPIYKVT
MSMEG_6105|M.smegmatis_MC2_155 EEAVDRVVGGPRRKSRIISEHEKKITAYHEGGHTLAAWAMPDIEPIYKVT
TH_0991|M.thermoresistible__bu EEAVDRVVGGPRRKSRLISELEKKITAYHEGGHTLAAWAMPDIEPIYKVT
MAB_0533|M.abscessus_ATCC_1997 EESVDRVVGGPRRKGRIISEHEKKITAYHEAGHTLAAWAMPDIERVYKVT
**:****:******.*:*** *********.************: :****
Rv3610c|M.tuberculosis_H37Rv ILARGRTGGHAVAVPEEDKGLRTRSEMIAQLVFAMGGRAAEELVFREPTT
Mb3640c|M.bovis_AF2122/97 ILARGRTGGHAVAVPEEDKGLRTRSEMIAQLVFAMGGRAAEELVFREPTT
MLBr_00222|M.leprae_Br4923 ILARGRTGGHAVAVPEEDKGLRTRSEMIAQLVFAMGGRAAEELVFREPTT
MAV_0542|M.avium_104 ILARGRTGGHAVAVPEEDKGLRTRSEMIAQLVFAMGGRAAEELVFREPTT
MMAR_5113|M.marinum_M ILARGRTGGHAVAVPEEDKGLRTRSEMIAQLVFAMGGRAAEELVFREPTT
MUL_4192|M.ulcerans_Agy99 ILARGRTGGHAVAVPEEDKGLRTRSEMIAQLVFAMGGRAAEELVFREPTT
Mflv_1416|M.gilvum_PYR-GCK ILARGRTGGHAMAVPEDDKGLMTRSEMIARLVFAMGGRAAEELVFREPTT
Mvan_5372|M.vanbaalenii_PYR-1 ILARGRTGGHAMSVPEDDKGLMTRSEMIARLVFAMGGRAAEELVFREPTT
MSMEG_6105|M.smegmatis_MC2_155 ILARGRTGGHAVAVPEDDKGLMTRSEMIARLVFAMGGRAAEELVFREPTT
TH_0991|M.thermoresistible__bu ILARGRTGGHAVAVPEDDKGLMTRSEMIARLVFAMGGRAAEELVFREPTT
MAB_0533|M.abscessus_ATCC_1997 ILARGRTGGHAIAVPEDDKGLATRSEMIAQLVFAMGGRAAEELVFREPTT
***********::***:**** *******:********************
Rv3610c|M.tuberculosis_H37Rv GAVSDIEQATKIARSMVTEFGMSSKLGAVKYGSEHGDPFLGRTMGTQPDY
Mb3640c|M.bovis_AF2122/97 GAVSDIEQATKIARSMVTEFGMSSKLGAVKYGSEHGDPFLGRTMGTQPDY
MLBr_00222|M.leprae_Br4923 GAVSDIEKATKIARSMVTEFGMSSKLGAVRYGSEHGDPFLGRTMGTQADY
MAV_0542|M.avium_104 GAVSDIEQATKIARAMVTEFGMSSKLGAVKYGSEHGDPFLGRTMGTQADY
MMAR_5113|M.marinum_M GAVSDIEQATKVARAMVTEYGMSARLGAVKYGTEHGDPFLGRSMGTQSDY
MUL_4192|M.ulcerans_Agy99 GAVSDIEQATKVARAMVTEYGMSARLGAVKYGTEHGDPFLGRSMGTQSDY
Mflv_1416|M.gilvum_PYR-GCK GASSDIDQATKIARAMVTEYGMSSKLGAVRYGSEHGDPFLGRTMGNQADY
Mvan_5372|M.vanbaalenii_PYR-1 GAVSDIEQATKIARAMVTEYGMSSKLGAVRYGTEHGDPFLGRTMGTQADY
MSMEG_6105|M.smegmatis_MC2_155 GAVSDIEQATKIARAMVTEYGMSSKLGAVRYGTEHGDPFLGRTMGTQADY
TH_0991|M.thermoresistible__bu GAVSDIEQATKIARAMVTEYGMSSKLGAVRYGTEHGDPFLGRTMGTHADY
MAB_0533|M.abscessus_ATCC_1997 GAVSDIEQATKKARAMVTEFGMSAKLGAVRYGTEHGDPFLGRTMGTQADY
** ***::*** **:****:***::****:**:*********:**.:.**
Rv3610c|M.tuberculosis_H37Rv SHEVAREIDEEVRKLIEAAHTEAWEILTEYRDVLDTLAGELLEKETLHRP
Mb3640c|M.bovis_AF2122/97 SHEVAREIDEEVRKLIEAAHTEAWEILTEYRDVLDTLAGELLEKETLHRP
MLBr_00222|M.leprae_Br4923 SHEVARDIDDEVRKLIEAAHTEAWEILTEYRDVLDTLAGELLEKETLHRP
MAV_0542|M.avium_104 SHEVARDIDDEIRKLIEAAHTEAWEILTEYRDILDTLAGQLLEKETLHRA
MMAR_5113|M.marinum_M SHEVAREIDEEVRKLIEAAHTEAWEILTEYRDVLDTLAGQLLEKETLHRP
MUL_4192|M.ulcerans_Agy99 SHEVAREIDEEVRKLIEAAHTEAWEILTEYRDVLDTLAGQLLEKETLHRP
Mflv_1416|M.gilvum_PYR-GCK SHEVAQIIDDEIRKLIEAAHTEAWEILTEYRDVLDTLAGELLEKETLHRV
Mvan_5372|M.vanbaalenii_PYR-1 SHEVAQIIDDEVRKLIEAAHTEAWEILTEYRDVLDTLAGELLEKETLHRV
MSMEG_6105|M.smegmatis_MC2_155 SHEVAQIIDDEVRKLIEAAHTEAWEILTEYRDVLDILAGELLEKETLHRA
TH_0991|M.thermoresistible__bu SHEVARDIDDEIRKLIEAAHTEAWEILTEYRDVLDVLAGELLEKETLHRR
MAB_0533|M.abscessus_ATCC_1997 SHEVAREIDEEVRNLIEAAHTEAWAILTEYRDVLDTLAGALLEKETVVRK
*****: **:*:*:********** *******:** *** ******: *
Rv3610c|M.tuberculosis_H37Rv ELESIFADVEKRPRLTMFDDFGGRIPSDKPPIKTPGELAIERGEPWPQPV
Mb3640c|M.bovis_AF2122/97 ELESIFADVEKRPRLTMFDDFGGRIPSDKPPIKTPGELAIERGEPWPQPV
MLBr_00222|M.leprae_Br4923 ELEGIFASVEKRPRLTMFDDFGGRIPSDKPPIKTPGELAIERGEPWPQPV
MAV_0542|M.avium_104 ELESIFAGVEKRPRLTMFDDFGGRIPSDKPPIKTPGELAIERGEPWPPPA
MMAR_5113|M.marinum_M ELESIFSDVEKRPRLTMFDDFGGRIPSDKPPIKTPGELAIERGEPWPQPM
MUL_4192|M.ulcerans_Agy99 ELESIFSDVEKRPRLTMFDDFGGRIPSDKPPIKTPGELAIERGEPWPQPM
Mflv_1416|M.gilvum_PYR-GCK ELEAIFGDVKKRPRLTMFDDFGGRVPSDKPPIKTPGELAIERGEPWPKPV
Mvan_5372|M.vanbaalenii_PYR-1 ELEAIFGDVKKRPRLTMFDDFGGRVPSDKPPIKTPGELAIERGEPWPKPL
MSMEG_6105|M.smegmatis_MC2_155 ELEAIFGDVKKRPRLTMFDDFGGRVPSDKPPIKTPGELAMERGEPWPPPV
TH_0991|M.thermoresistible__bu ELEVILADVKKRPRLTMFDDFGGRTPSDKPPIKTPGELAMERGEPWPKPE
MAB_0533|M.abscessus_ATCC_1997 ELEEIFSGVQRRPRLTMFDDFGGRIPSDKPPIKTPGELAIERGEPWPPPV
*** *:..*::************* **************:******* *
Rv3610c|M.tuberculosis_H37Rv PEPAFKAAIAQATQAAEAARSDAGQTGHGANGSPAGTHRSGDRQ---YGS
Mb3640c|M.bovis_AF2122/97 PEPAFKAAIAQATQAAEAARSDAGQTGHGANGSPAGTHRSGDRQ---YGS
MLBr_00222|M.leprae_Br4923 PEPAFKAAIARASQAAEASHQAAQSDTDGPPGQGANGSHAGDRQR-QHGP
MAV_0542|M.avium_104 PEPAFKAAIAKASEAAAAARLEADRNANGGNGSHAN--QGGNRQ----GP
MMAR_5113|M.marinum_M PEPAFKQAIAQATQAAQAA--QAAQSASEGTGRGPNGSRGADAN----PG
MUL_4192|M.ulcerans_Agy99 PEPAFKQAIAQAT--------QAAQSASEGTGRGPNGSRGADAN----PG
Mflv_1416|M.gilvum_PYR-GCK PEPAFKTAIAQASRAAAEAAPKNGGNGANGAH-APG--------------
Mvan_5372|M.vanbaalenii_PYR-1 PEPAFKTAIAQASRAAAEQAQKNGGNGSQASNGVPGG------------P
MSMEG_6105|M.smegmatis_MC2_155 PEPAFKAAIAAANREAEKRNGTNGAP-SNGAP-APAG------------P
TH_0991|M.thermoresistible__bu PEPAFKVAIAEATRAAEAASGQNGQNGHNGTPGPTGPNGTPSAPAQQPAP
MAB_0533|M.abscessus_ATCC_1997 PEPAFKKAIAEQAAAAAPANGVPPAPAGPHGHLPNGQGG-------QPVP
****** ***
Rv3610c|M.tuberculosis_H37Rv TQPDYGAPAGWHAPGWPPRSS--------HRPS---YSGEPAPTYPGQ--
Mb3640c|M.bovis_AF2122/97 TQPDYGAPAGWHAPGWPPRSS--------HRPS---YSGEPAPTYPGQ--
MLBr_00222|M.leprae_Br4923 TQPDYGAPPGWHAPGWPPQQPPDYWYPPEQQPSQSPYWPQPAPSYPGQAP
MAV_0542|M.avium_104 TQPDYGAPAGWRAPGWPPQQSQQP--NYWHPPAQPPYWQQPAHGYPGQQP
MMAR_5113|M.marinum_M SHPDYGAPAGWRAPGWPPPSE----------------RPQPSGYSPDQ--
MUL_4192|M.ulcerans_Agy99 SHPDYGAPAGWRAPGWPPPSE----------------RPQPSGYSPDQ--
Mflv_1416|M.gilvum_PYR-GCK --PDYGAPAGWHAPGWPPPPGGPQQQQPPQQ---QGYWYPGPPQQQGWQG
Mvan_5372|M.vanbaalenii_PYR-1 TQPDYGAPAGWHAPGWPPQAGGP---HPPQP---QGYWYP-PPPPSGWQG
MSMEG_6105|M.smegmatis_MC2_155 TQPDYGAPAGWHAPGWPPPQQSQQPGYQPQ----QGYWYP-PPHPSGWQ-
TH_0991|M.thermoresistible__bu VVPDYGAPAGWHAPGWPPQPTPQQQPPPYQQP--HGQWYP-PQQQPGWQ-
MAB_0533|M.abscessus_ATCC_1997 PGVNYGAPAGWHAPGWPPPGQAPHP-------------------QPGYQ-
:****.**:****** .
Rv3610c|M.tuberculosis_H37Rv -----PYP--------------------------TGQADPGSDESSAEQD
Mb3640c|M.bovis_AF2122/97 -----PYP--------------------------TGQADPGSDESSAEQD
MLBr_00222|M.leprae_Br4923 P----PYPS----------------YPPCPSYPPPGQSAPDAGKPPAQLD
MAV_0542|M.avium_104 YPGQQPYPGQPGGPAQSGPPQARPPYPPYPPYPPPGQPAPEGG-SPDRQD
MMAR_5113|M.marinum_M --------------------------------------PSDTDESSEQQD
MUL_4192|M.ulcerans_Agy99 --------------------------------------PSDTDESSEQQD
Mflv_1416|M.gilvum_PYR-GCK NAPTPAYPG----------------YPPYQPYPQPGQHAPHGGAPAPNPT
Mvan_5372|M.vanbaalenii_PYR-1 APQAPAYPG----------------YPPYQPHSQPGQPAPHGGAAAASPK
MSMEG_6105|M.smegmatis_MC2_155 --QPQPYP--------------------YQPYPHPGQQGTS--PEHGRPS
TH_0991|M.thermoresistible__bu -QQQQPYPSQ---------------YSPYQPYPQPSPPAPPGGAAPND--
MAB_0533|M.abscessus_ATCC_1997 -----PYPP--------------------QPYPYP----------VPTPH
Rv3610c|M.tuberculosis_H37Rv DEVSRTKP-AHG----------------------
Mb3640c|M.bovis_AF2122/97 DEVSRTKP-AHG----------------------
MLBr_00222|M.leprae_Br4923 EGVSPSNPPAHG----------------------
MAV_0542|M.avium_104 DDVSRSNPPAHG----------------------
MMAR_5113|M.marinum_M RDVSRSNPPAHG----------------------
MUL_4192|M.ulcerans_Agy99 RDVSRSNPPAHG----------------------
Mflv_1416|M.gilvum_PYR-GCK DDPGQDRGRENGHRDTERRDTDRGDTNRNGPANG
Mvan_5372|M.vanbaalenii_PYR-1 EDPGHEGGGENGRSAP--------------PSNG
MSMEG_6105|M.smegmatis_MC2_155 EQSGQDDGHTGDRSNP----------------HG
TH_0991|M.thermoresistible__bu QESGQDPNRTNPPGQG------------------
MAB_0533|M.abscessus_ATCC_1997 QQPGPAPDEGSESKG-------------------
.