For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RRLISVAYALMVATIVGLSAAGGWFYWDRVQTGGEASARALLPKLAMQEIPQVFGYDYQTVERSLTAVYP LLTPDYRQEFQKSANAQIIPEAKKREVVVQANVVGVGVMDAKRDCASVMVYLNRTVTDKTRQPLYDGSRL RVDFQRIDGKWLIAYITPI*
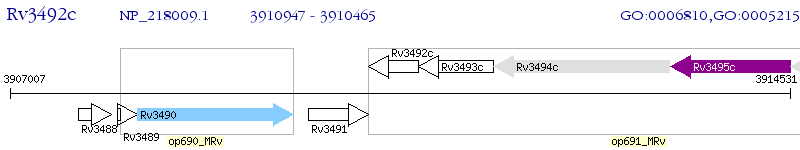
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3492c | - | - | 100% (160) | CONSERVED HYPOTHETICAL MCE ASSOCIATED PROTEIN |
| M. tuberculosis H37Rv | Rv1973 | - | 1e-12 | 25.32% (154) | MCE associated membrane protein |
| M. tuberculosis H37Rv | Rv1362c | - | 3e-09 | 23.38% (154) | hypothetical protein Rv1362c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3522c | - | 8e-89 | 100.00% (160) | Mce associated protein |
| M. gilvum PYR-GCK | Mflv_1563 | - | 4e-54 | 62.99% (154) | hypothetical protein Mflv_1563 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4146c | - | 2e-44 | 51.88% (160) | MCE-family protein |
| M. marinum M | MMAR_4980 | - | 8e-69 | 78.62% (159) | MCE-associated protein |
| M. avium 104 | MAV_0664 | - | 4e-65 | 76.10% (159) | hypothetical protein MAV_0664 |
| M. smegmatis MC2 155 | MSMEG_5893 | - | 1e-53 | 59.49% (158) | hypothetical protein MSMEG_5893 |
| M. thermoresistible (build 8) | TH_2240 | - | 1e-51 | 61.39% (158) | CONSERVED HYPOTHETICAL MCE ASSOCIATED PROTEIN |
| M. ulcerans Agy99 | MUL_4055 | - | 4e-69 | 78.62% (159) | MCE-associated protein |
| M. vanbaalenii PYR-1 | Mvan_5195 | - | 8e-53 | 59.49% (158) | hypothetical protein Mvan_5195 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3492c|M.tuberculosis_H37Rv ------MRRLISVAYALMVATIVGLSAAGGWFYWDRVQTGGEASARALLP
Mb3522c|M.bovis_AF2122/97 ------MRRLISVAYALMVATIVGLSAAGGWFYWDRVQTGGEASARALLP
MMAR_4980|M.marinum_M MRLRRWQRWVITVAASMLAIVIVGLAALGGWFYWDRVEARGEAAARAVLP
MUL_4055|M.ulcerans_Agy99 MRLRRWQRWVITVAASMLAVVIVGLAALGGWFYWDRVEARGEAAARAVLP
MAV_0664|M.avium_104 ---MRWRRRLLVAAGYLLAVAFVGLSAVAGAMYWDRVQTRGEQAARAVLP
Mflv_1563|M.gilvum_PYR-GCK --MRDGLAKVIGAVAIVLATGFVALSAVGGWLYWKGVELDGEQQARAELP
Mvan_5195|M.vanbaalenii_PYR-1 --MRDWPAKLIAAIGVLLATGFVALSAVGGALYWNRVELNGEEQARAELP
TH_2240|M.thermoresistible__bu --MSRWAVRLVAGGAILLAVAFVVLSAVGGGLYWNRVELRAEQLTRETLG
MSMEG_5893|M.smegmatis_MC2_155 --MR---SKLVGALVGVLAVAFIALSALGGAMYWSRVQQRGQEATRAELP
MAB_4146c|M.abscessus_ATCC_199 ------MRWLARIFVAVTVLVVVGLSAAAGGMYWHRVDTVASRAAGETLI
: : . .: *:* .* :** *: .. : *
Rv3492c|M.tuberculosis_H37Rv KLAMQEIPQVFGYDYQTVERSLTAVYPLLTPDYRQEFQKSANAQIIPEAK
Mb3522c|M.bovis_AF2122/97 KLAMQEIPQVFGYDYQTVERSLTAVYPLLTPDYRQEFQKSANAQIIPEAK
MMAR_4980|M.marinum_M GLAAKDIPLVFGYDYQTVERSLNAAYPLLTPAYREEFEKSANAQIIPEAK
MUL_4055|M.ulcerans_Agy99 GLAAKDIPLVFGYDYQTVERSLNAAYPLLTPAYREEFEKSANAQIIPEAK
MAV_0664|M.avium_104 GLAAKEIPEVFAYDYQTVERSLSAAYPLLTPDYRAEFQKSVNNQIIPEAK
Mflv_1563|M.gilvum_PYR-GCK ALAEEQIPRVFGFDYQTVERSFAEVYPMLTPEYRQEFQDRVTNEIIPQAR
Mvan_5195|M.vanbaalenii_PYR-1 SLAAQQIPLVFGFDYQTVERTFAEVYPLLTPEYRKEFQDRVTNEIIPQAR
TH_2240|M.thermoresistible__bu PLAEEQIPAVFGYDYQTVERSLTEAYDLLTPSYREEFRDRATKDIIPQAR
MSMEG_5893|M.smegmatis_MC2_155 SLAIEQIPRVFGYDYQTVERSLMETYPLLTPGFRQQFESDAQQKVIPEAR
MAB_4146c|M.abscessus_ATCC_199 KQAGKQIGSIFGYDYQTVERSLDEQYAGLTPEYRREFEDKANREIIPQAK
* ::* :*.:*******:: * *** :* :*.. . .:**:*:
Rv3492c|M.tuberculosis_H37Rv KREVVVQANVVGVGVMDAKRDCASVMVYLNRTVTDKTRQPLYDGSRLRVD
Mb3522c|M.bovis_AF2122/97 KREVVVQANVVGVGVMDAKRDCASVMVYLNRTVTDKTRQPLYDGSRLRVD
MMAR_4980|M.marinum_M KREVVVQANVVGVGVMAAKRNSASVMVYLNRTVTDKSRQPLYDGSRLRVD
MUL_4055|M.ulcerans_Agy99 KREVVVQANVVGVGVMAAKRNSASVMVYLNRTVTDKSRQPLYDGSRLRVD
MAV_0664|M.avium_104 KREVVVQANVVGVGVMSAQRDSASVMVYMNRTVTDKSRQPLYDGSRLRVD
Mflv_1563|M.gilvum_PYR-GCK DRELISQAHTVGTGVLDAHRTSASVMVYMNRTVTDKSKESVYDGSRLRVD
Mvan_5195|M.vanbaalenii_PYR-1 DRQLISQAHSVGAGILDAHRTSASVMVYMNRTVTDKAKESVYDGSRLRVD
TH_2240|M.thermoresistible__bu EREVVSQANVVGVGVLAAQRNSGSVMVYMNRTVTDKSKQSVYDGSRLRVD
MSMEG_5893|M.smegmatis_MC2_155 KRELVVQINIVGVGVEAAQRDTGSVLVYMNRTVTDKSRQPLYDGSRLRVE
MAB_4146c|M.abscessus_ATCC_199 QRKIVSQTSIVGTGIIAAHRDSAQVVVYMNRTITDQSRTPVYDSSSIQVD
.*::: * **.*: *:* ..*:**:***:**::: .:**.* ::*:
Rv3492c|M.tuberculosis_H37Rv FQRIDGKWLIAYITPI
Mb3522c|M.bovis_AF2122/97 FQRIDGKWLIAYITPI
MMAR_4980|M.marinum_M YKRIGGKWLIAYITPI
MUL_4055|M.ulcerans_Agy99 YKRIGGKWLIAYITPI
MAV_0664|M.avium_104 FKKIGKQWLIAYITPI
Mflv_1563|M.gilvum_PYR-GCK YKKVDGKWLIAYITPI
Mvan_5195|M.vanbaalenii_PYR-1 YKKVDGKWLIAYITPI
TH_2240|M.thermoresistible__bu YEKIDGKWLIDYITPI
MSMEG_5893|M.smegmatis_MC2_155 YRKVDGKWLIDAIKPI
MAB_4146c|M.abscessus_ATCC_199 YTRAGDKWLISYITPL
: : . :*** *.*: