For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTVLGIETSCDETGVGIARLDPDGTVTLLADEVASSVDEHVRFGGVVPEIASRAHLEALGPAMRRALAAA GLKQPDIVAATIGPGLAGALLVGVAAAKAYSAAWGVPFYAVNHLGGHLAADVYEHGPLPECVALLVSGGH THLLHVRSLGEPIIELGSTVDDAAGEAYDKVARLLGLGYPGGKALDDLARTGDRDAIVFPRGMSGPADDR YAFSFSGLKTAVARYVESHAADPGFRTADIAAGFQEAVADVLTMKAVRAATALGVSTLLIAGGVAANSRL RELATQRCGEAGRTLRIPSPRLCTDNGAMIAAFAAQLVAAGAPPSPLDVPSDPGLPVMQGQVR*
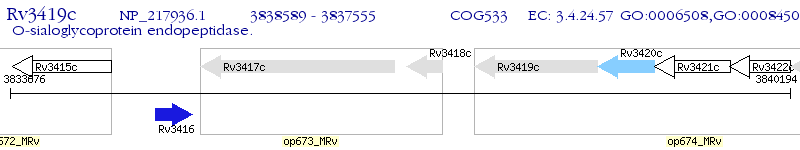
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3419c | gcp | - | 100% (344) | O-sialoglycoprotein endopeptidase |
| M. tuberculosis H37Rv | Rv3421c | - | 4e-07 | 31.50% (127) | hypothetical protein Rv3421c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3453c | gcp | 0.0 | 100.00% (344) | putative DNA-binding/iron metalloprotein/AP endonuclease |
| M. gilvum PYR-GCK | Mflv_4929 | - | 1e-160 | 83.53% (340) | O-sialoglycoprotein endopeptidase |
| M. leprae Br4923 | MLBr_00379 | gcp | 1e-164 | 86.09% (345) | putative DNA-binding/iron metalloprotein/AP endonuclease |
| M. abscessus ATCC 19977 | MAB_3734c | - | 1e-153 | 78.43% (343) | putative DNA-binding/iron metalloprotein/AP endonuclease |
| M. marinum M | MMAR_1124 | gcp | 1e-170 | 87.72% (342) | O-sialoglycoprotein endopeptidase Gcp (glycoprotease) |
| M. avium 104 | MAV_4367 | - | 1e-168 | 86.59% (343) | O-sialoglycoprotein endopeptidase |
| M. smegmatis MC2 155 | MSMEG_1580 | - | 1e-162 | 84.41% (340) | O-sialoglycoprotein endopeptidase |
| M. thermoresistible (build 8) | TH_1689 | gcp | 1e-161 | 84.41% (340) | PROBABLE O-SIALOGLYCOPROTEIN ENDOPEPTIDASE GCP |
| M. ulcerans Agy99 | MUL_0884 | gcp | 1e-169 | 87.43% (342) | putative DNA-binding/iron metalloprotein/AP endonuclease |
| M. vanbaalenii PYR-1 | Mvan_1489 | - | 1e-161 | 83.82% (340) | O-sialoglycoprotein endopeptidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4929|M.gilvum_PYR-GCK --------MIILAIESSCDETGVGIAELGDDGSVTLLADEVASSVDEHAR
Mvan_1489|M.vanbaalenii_PYR-1 --------MIILAVESSCDETGVGIAELGDDGTVTLLADEVASSVDEHAR
TH_1689|M.thermoresistible__bu --------MIILAIESSCDETGVGIAELAPDGSVTLLADEVASSVDEHAR
MSMEG_1580|M.smegmatis_MC2_155 --------MIILAIESSCDETGVGIADLRDDGTVTLLADEVASSVDEHAR
Rv3419c|M.tuberculosis_H37Rv -------MTTVLGIETSCDETGVGIARLDPDGTVTLLADEVASSVDEHVR
Mb3453c|M.bovis_AF2122/97 -------MTTVLGIETSCDETGVGIARLDPDGTVTLLADEVASSVDEHVR
MMAR_1124|M.marinum_M -----MPGTVILAIETSCDETGVGITRLEADGTLTLLADEVASSVDEHVR
MUL_0884|M.ulcerans_Agy99 -----MPGTIILAIETSCDETGVGITRLEADGTLTLLADEVASSVDEHVR
MLBr_00379|M.leprae_Br4923 MTISAVPGTIILAIETSCDETGVGIACLDDYGTVTLLADEVASSVDEQAR
MAV_4367|M.avium_104 -------MTTILAIETSCDETGVGIARLDADGAVTLLADEVASSVDEHVR
MAB_3734c|M.abscessus_ATCC_199 -------MTVILAIESSCDETGVGIAELTPDGSVVLLADEVASSVEEHAR
:*.:*:*********: * *::.**********:*:.*
Mflv_4929|M.gilvum_PYR-GCK FGGVVPEIASRAHLEALGPTMRRALATAGVD---KPDVVAATIGPGLAGA
Mvan_1489|M.vanbaalenii_PYR-1 FGGVVPEIASRAHLEALGPTMRRALATAGVS---KPDVVAATIGPGLAGA
TH_1689|M.thermoresistible__bu FGGVVPEIASRAHLEALQPTMRRALAAAGVR---RPDVVAATIGPGLAGA
MSMEG_1580|M.smegmatis_MC2_155 FGGVVPEIASRAHLEALGPTMRRALDAAGIE---RPDVVAATIGPGLAGA
Rv3419c|M.tuberculosis_H37Rv FGGVVPEIASRAHLEALGPAMRRALAAAGLK---QPDIVAATIGPGLAGA
Mb3453c|M.bovis_AF2122/97 FGGVVPEIASRAHLEALGPAMRRALAAAGLK---QPDIVAATIGPGLAGA
MMAR_1124|M.marinum_M FGGVVPEIASRAHLEALGPAMRRALTTAGIV---RPDVVATTIGPGLAGA
MUL_0884|M.ulcerans_Agy99 FGGVVPEIASRAHLEALGPAMRRALTTAGIV---RPDVVATTIGPGLAGA
MLBr_00379|M.leprae_Br4923 FGGVVPEIASRAHLEALGPTIRCALAAAGLTGSAKPDVVAATIGPGLAGA
MAV_4367|M.avium_104 FGGVVPEIASRAHLEALGPTMRRALSAAGVT---KPDVVAATIGPGLAGA
MAB_3734c|M.abscessus_ATCC_199 FGGVVPEIASRAHLEALGVTARRALDLAGVQ---RPDVVAATIGPGLAGA
***************** : * ** **: :**:**:*********
Mflv_4929|M.gilvum_PYR-GCK LLVGVAAAKAYAAAWQVPFYAVNHLGGHLAADVFDHGPLPESIGLLVSGG
Mvan_1489|M.vanbaalenii_PYR-1 LLVGVAAAKAYSAAWQVPFYAVNHLGGHLAADVFDHGPLPESIGLLVSGG
TH_1689|M.thermoresistible__bu LLVGVAAAKAYAAAWQVPFYAVNHLGGHLAADVYEHGPLPESVGLLVSGG
MSMEG_1580|M.smegmatis_MC2_155 LLVGVAAAKAYAAGWGVPFYGVNHLGGHLAADVYDHGPLPESVGLLVSGG
Rv3419c|M.tuberculosis_H37Rv LLVGVAAAKAYSAAWGVPFYAVNHLGGHLAADVYEHGPLPECVALLVSGG
Mb3453c|M.bovis_AF2122/97 LLVGVAAAKAYSAAWGVPFYAVNHLGGHLAADVYEHGPLPECVALLVSGG
MMAR_1124|M.marinum_M LLVGVAAAKAYAAAWGVPFYAVNHLGGHLAADVYEHGPLPECVALLVSGG
MUL_0884|M.ulcerans_Agy99 LLVGVAAAKAYAAAWGVPFYAVNHLGGHLAADVYEHGPLPECVALLVSGG
MLBr_00379|M.leprae_Br4923 LLVGVAAAKAYSAAWGVPFYAVNHLGGHLAADVYEHGPLPECVALLVSGG
MAV_4367|M.avium_104 LLVGVAAAKAYSAAWGVPFYAVNHLGGHLAADVYQHGPLPECVALLVSGG
MAB_3734c|M.abscessus_ATCC_199 LLVGVSAAKGLALAWGVPFYGVNHLGGHIAADVYENGPLPECVALLVSGG
*****:***. : .* ****.*******:****:::*****.:.******
Mflv_4929|M.gilvum_PYR-GCK HTHLLHVRSLGEPIIELGSTVDDAAGEAYDKIARLLGLGYPGGRVLDELA
Mvan_1489|M.vanbaalenii_PYR-1 HTHLLHVRSLGEPIVELGSTVDDAAGEAYDKIARLLGLGYPGGRVLDELA
TH_1689|M.thermoresistible__bu HTHLLHVRSLGEPIIELGSTVDDAAGEAYDKVARLLGLGYPGGRVLDELA
MSMEG_1580|M.smegmatis_MC2_155 HTHLLHVRSLGEPIIELGSTVDDAAGEAYDKVARLLGLGYPGGRVLDELA
Rv3419c|M.tuberculosis_H37Rv HTHLLHVRSLGEPIIELGSTVDDAAGEAYDKVARLLGLGYPGGKALDDLA
Mb3453c|M.bovis_AF2122/97 HTHLLHVRSLGEPIIELGSTVDDAAGEAYDKVARLLGLGYPGGKALDDLA
MMAR_1124|M.marinum_M HTHLLHVRSLGEPMVELGATVDDAAGEAYDKVARLLGLGYPGGKVLDDLA
MUL_0884|M.ulcerans_Agy99 HTHLLHVRSLGEPMVELGATVDDAAGEAYDKVARLLGLGYPGGKVLDDLA
MLBr_00379|M.leprae_Br4923 HTHLLQVRSLGAPIVELGSTVDDAAGEAYDKVARLLGLGYPGGKVLDDLA
MAV_4367|M.avium_104 HTHLLQVRSLGDPIVELGSTVDDAAGEAYDKVARLLGLGYPGGRVLDELA
MAB_3734c|M.abscessus_ATCC_199 HTSLLHVRSLAEPIEELGATVDDAAGEAYDKVARLLGLGYPGGKVLDELA
** **:****. *: ***:************:***********:.**:**
Mflv_4929|M.gilvum_PYR-GCK REGDPKAITFPRGMTGPRDEPYAFSFSGLKTAVARYVESH---PEASQAD
Mvan_1489|M.vanbaalenii_PYR-1 REGDPAAITFPRGMTGPRDDPYAFSFSGLKTAVARYVESH---PEASQAD
TH_1689|M.thermoresistible__bu RQGDSTAITFPRGMTGPRDAPYSFSFSGLKTAVARYVESH---PDAAPAD
MSMEG_1580|M.smegmatis_MC2_155 RTGDRDAIVFPRGMTGPRDDPYTFSFSGLKTAVARYVEAH---PEASHAD
Rv3419c|M.tuberculosis_H37Rv RTGDRDAIVFPRGMSGPADDRYAFSFSGLKTAVARYVESHAADPGFRTAD
Mb3453c|M.bovis_AF2122/97 RTGDRDAIVFPRGMSGPADDRYAFSFSGLKTAVARYVESHAADPGFRTAD
MMAR_1124|M.marinum_M RSGDPDAIVFPRGMTGPSDDPCAFSFSGLKTAVARYVEAH---PDFRPAD
MUL_0884|M.ulcerans_Agy99 RSGDPDAIVFPRGMTGPSDDPCAFSFSGLKTAVACYVEAH---PDFRPAD
MLBr_00379|M.leprae_Br4923 RTGDRDAIVFPRGMTGPADDLNAFSFSGLKTAVARYVESH---PDALPAD
MAV_4367|M.avium_104 RTGDREAIVFPRGMTGPRDAPYAFSFSGLKTAVARYLESH---PDAVNAD
MAB_3734c|M.abscessus_ATCC_199 QQGDSSAVPFPRGMTGPRDARHSFSFSGLKTAVARYLEKHGVEADFSPAD
: ** *: *****:** * :*********** *:* * . **
Mflv_4929|M.gilvum_PYR-GCK VAAGFQEAVADVLTAKAVRAAGDLGVSTLLIAGGVAANSRLRELAAQRCE
Mvan_1489|M.vanbaalenii_PYR-1 VAAGFQEAVADVLTAKAVRAAKDLGVSTLLIAGGVAANSRLRELAGQRCA
TH_1689|M.thermoresistible__bu IAAGFQEAVADVLTAKAVRACTDLGVSTLLIAGGVAANSRLRELAAERCA
MSMEG_1580|M.smegmatis_MC2_155 VAAGFQESVADVLTAKAVRAATDLGVSTLLIAGGVAANSRLRELAEERCA
Rv3419c|M.tuberculosis_H37Rv IAAGFQEAVADVLTMKAVRAATALGVSTLLIAGGVAANSRLRELATQRCG
Mb3453c|M.bovis_AF2122/97 IAAGFQEAVADVLTMKAVRAATALGVSTLLIAGGVAANSRLRELATQRCG
MMAR_1124|M.marinum_M VAAGFQEAVADVLTRKAVRAATGLGVSTLLIAGGVAANSRLRELATQRCS
MUL_0884|M.ulcerans_Agy99 VAAGFQEAVADVLTRKAVRAATGLGVSTLLIAGGVAANSRLRELATQRCS
MLBr_00379|M.leprae_Br4923 VAAGFQEAVADVLTMKAVRAATGLGVSTLLIVGGVAANSRLRELAAQRCA
MAV_4367|M.avium_104 VAAGFQEAVADVLTRKAVRAATDLGVTTLLIAGGVAANSRLRELAEQRCA
MAB_3734c|M.abscessus_ATCC_199 VAAGFQEAVADVLTMKAVRAATDLGVSTLLIAGGVAANSRVRALAEQRCA
:******:****** *****. ***:****.********:* ** :**
Mflv_4929|M.gilvum_PYR-GCK AAGMTLRIPRPRLCTDNGAMIASFAAHLIAAGAAPSPLDAASDPGLPVVQ
Mvan_1489|M.vanbaalenii_PYR-1 DAGMTLRIPRPRLCTDNGAMIASFAAHLIAAGARPSPLDAASDPGLPVVQ
TH_1689|M.thermoresistible__bu EAALTLRIPRPRLCTDNGAMIASYAAHLIAAGAAPSPLDCASDPGLPVVQ
MSMEG_1580|M.smegmatis_MC2_155 AAGLTLRVPRPRLCTDNGAMIASFAAHLIAAGAQPSPLDAASDPGLPVVK
Rv3419c|M.tuberculosis_H37Rv EAGRTLRIPSPRLCTDNGAMIAAFAAQLVAAGAPPSPLDVPSDPGLPVMQ
Mb3453c|M.bovis_AF2122/97 EAGRTLRIPSPRLCTDNGAMIAAFAAQLVAAGAPPSPLDVPSDPGLPVMQ
MMAR_1124|M.marinum_M EAGLTLRIPRPRLCTDNGAMIAAFAAHLVAAKAAPSPLDVPSDPGLPVVK
MUL_0884|M.ulcerans_Agy99 EAGLTLRIPRPRLCTDNGAMIAAFAAHLVAAKAAPSPLDVPSDPGLPVVK
MLBr_00379|M.leprae_Br4923 AAGLMLRIPGPRFCTDNGAMIAAFAAHLLAAAAPPSPLDVPSDPGLPVVK
MAV_4367|M.avium_104 AAGLTLRIPRPGLCTDNGAMIASFAAHLVAAGAPPSPLDVPTDPGLPVVK
MAB_3734c|M.abscessus_ATCC_199 ETGLTLRVPPLRLCTDNGAMIASFAAHLVAAGAQPSSLQAAADPGLPVVK
:. **:* :*********::**:*:** * **.*: .:******::
Mflv_4929|M.gilvum_PYR-GCK GQVA
Mvan_1489|M.vanbaalenii_PYR-1 GQVA
TH_1689|M.thermoresistible__bu GQVV
MSMEG_1580|M.smegmatis_MC2_155 GQVA
Rv3419c|M.tuberculosis_H37Rv GQVR
Mb3453c|M.bovis_AF2122/97 GQVR
MMAR_1124|M.marinum_M GQVG
MUL_0884|M.ulcerans_Agy99 GQVG
MLBr_00379|M.leprae_Br4923 RQIN
MAV_4367|M.avium_104 AQVS
MAB_3734c|M.abscessus_ATCC_199 GQVH
*: