For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SYPENVLAAGEQVVLHRHPHWNRLIWPVVVLVLLTGLAAFGSGFVNSTPWQQIAKNVIHAVIWGIWLVIV GWLTLWPFLSWLTTHFVVTNRRVMFRHGVLTRSGIDIPLARINSVEFRDRIFERIFRTGTLIIESASQDP LEFYNIPRLREVHALLYHEVFDTLGSDESPS*
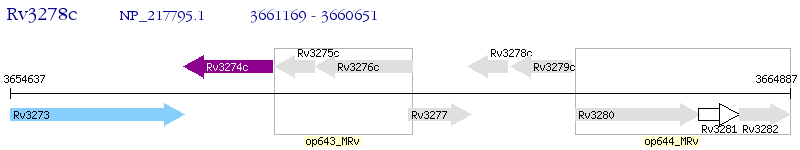
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3278c | - | - | 100% (172) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. tuberculosis H37Rv | Rv1226c | - | 4e-05 | 25.00% (96) | hypothetical protein Rv1226c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3306c | - | 1e-100 | 99.42% (172) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_4749 | - | 3e-70 | 68.60% (172) | membrane-flanked domain-containing protein |
| M. leprae Br4923 | MLBr_00733 | - | 3e-87 | 83.14% (172) | hypothetical protein MLBr_00733 |
| M. abscessus ATCC 19977 | MAB_3625c | - | 2e-71 | 71.51% (172) | hypothetical protein MAB_3625c |
| M. marinum M | MMAR_1258 | - | 8e-92 | 88.95% (172) | hypothetical protein MMAR_1258 |
| M. avium 104 | MAV_4246 | - | 1e-84 | 83.14% (172) | hypothetical protein MAV_4246 |
| M. smegmatis MC2 155 | MSMEG_1818 | - | 2e-74 | 74.42% (172) | membrane flanked domain-containing protein |
| M. thermoresistible (build 8) | TH_0887 | - | 2e-72 | 70.35% (172) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_2630 | - | 9e-92 | 88.95% (172) | hypothetical protein MUL_2630 |
| M. vanbaalenii PYR-1 | Mvan_1707 | - | 1e-70 | 68.02% (172) | membrane-flanked domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1818|M.smegmatis_MC2_155 ---------MGYPDNVLAGDEQVVLHRHPHWKRLIGPVFILIVATALASF
TH_0887|M.thermoresistible__bu ---------VGYPDNVLASDEQVVLHRHPHWKALVAPVLTLLVASAVAAF
Mflv_4749|M.gilvum_PYR-GCK MLNPQYSAAVGYPENVLAKDEHVVLHRHPHWGRLVLPAVFLIIASAAAAF
Mvan_1707|M.vanbaalenii_PYR-1 ---------MGYPENVLATDEHVVLHRHPHWGRLTIPALILIIASAAAAF
MAB_3625c|M.abscessus_ATCC_199 ---------MGYPENVLADDEQVVLHRHPHWKRLIGPALVLILSTGIAAF
Rv3278c|M.tuberculosis_H37Rv ---------MSYPENVLAAGEQVVLHRHPHWNRLIWPVVVLVLLTGLAAF
Mb3306c|M.bovis_AF2122/97 ---------MSYPENVLAAGEQVVLHRHPHWNRLIWPVVVLVLLTGLAAF
MMAR_1258|M.marinum_M ---------MGYPDNAMAAGEQVVLHRHPHWTRLVWPVVVLILITGLAAL
MUL_2630|M.ulcerans_Agy99 ---------MGYPDNAMAAGEQVVLHRHPHWARLVWPVVVLILITGLAAL
MLBr_00733|M.leprae_Br4923 ---------MRYPGNTLVAGEQVVLHRHPHWKRLIWPAVVLILATGLVSF
MAV_4246|M.avium_104 ---------MGYPDNVLAAGEHVIVHRHPHWKRLIGPVLVLILGTGLAAF
: ** *.:. .*:*::****** * *.. *:: :. .::
MSMEG_1818|M.smegmatis_MC2_155 GAAYVNTLHWDPTATLVVSLVILAIWLVIVGWLTLWPFLTWLTTHFVITD
TH_0887|M.thermoresistible__bu AAGWVNTTDWDGTAKNVLYIVIGAIWLVVIGWLTLWPFLNWLTTHFVITD
Mflv_4749|M.gilvum_PYR-GCK IAGYVNTLNWEQAAKTIVFGVIAAIWLVLVGWLAVWPFLNWWTTHFVITD
Mvan_1707|M.vanbaalenii_PYR-1 LAGYVNTLNWEQNAKTIVFAVIAAVWLILVGWLSIWPFLNWWTTHFVITD
MAB_3625c|M.abscessus_ATCC_199 VAAMVDNTDWQSTAKNVVMIAIGVVWLVLVGWLSLWPFLNWLTTHFVITD
Rv3278c|M.tuberculosis_H37Rv GSGFVNSTPWQQIAKNVIHAVIWGIWLVIVGWLTLWPFLSWLTTHFVVTN
Mb3306c|M.bovis_AF2122/97 GSGFVNSTPWQQIAKNVIHAVIWGIWLVIVGWLTLWPFLSWLTTHFVVTN
MMAR_1258|M.marinum_M GSGFVNSTQWQQLAKNIIHGVIWGIWLVIVGWLTLWPFLSWLTTHFVVTN
MUL_2630|M.ulcerans_Agy99 GSGFVNSTQWQQLAKNIIHGVIWGIWLVIVGWLTLWPFLSWLTTHFVVTN
MLBr_00733|M.leprae_Br4923 GSGYVNSTHWAQVAKNVIYGVLWGVWLVIVGWLTLWPFLNWLTTHFVVTN
MAV_4246|M.avium_104 GSGYLDSTHFEQLAKNIIHGVIWGIWLVIVGWLTLWPFFGWLTTHFVVTN
:. ::. : *. :: .: :**:::***::***: * *****:*:
MSMEG_1818|M.smegmatis_MC2_155 RRVMFRHGLLTRSGIDIPLARINSVEFRHGLSDRILRTGTLIIESASQDP
TH_0887|M.thermoresistible__bu RRVMFRYGVLTRAGIDIPLARINSVEFRHNLMDRILRTGTLIIESAAQDP
Mflv_4749|M.gilvum_PYR-GCK RRVMYRHGLITRSGIDIPLARINSVEFRHGLVDRIFRTGTLIIESAAQDP
Mvan_1707|M.vanbaalenii_PYR-1 RRVMYRHGLLTRSGIDIPLARINSVEFRHGLVDRMFRTGTLIIESASQDP
MAB_3625c|M.abscessus_ATCC_199 RRVMFRHGLLSRSGIDIPLARINSVEFRHGLVDRILRTGTLIIESASQDP
Rv3278c|M.tuberculosis_H37Rv RRVMFRHGVLTRSGIDIPLARINSVEFRDRIFERIFRTGTLIIESASQDP
Mb3306c|M.bovis_AF2122/97 RRVMFRHGVLTRSGIDIPLARINSVEFRDRIFERIFRTGTLIIESASQDP
MMAR_1258|M.marinum_M RRVMYRQGVLTRSGIDIPLARINSVEFRDRIFERMLRTGTLIIESASQDP
MUL_2630|M.ulcerans_Agy99 RRVMYRQGVLTRSGIDIPLARINSVEFRDRIFERMLRTGTLIIESASQDP
MLBr_00733|M.leprae_Br4923 RRVMFRQGTLTRSGVDIPLARINSVEFRDRLFERMFRTGTLIIESASQDP
MAV_4246|M.avium_104 RRVMFRHGVATRTGIDIPLARINSVEFRDRITERMLRTGTLIIESASQDP
****:* * :*:*:*************. : :*::**********:***
MSMEG_1818|M.smegmatis_MC2_155 LEFYDIPRVEQVHALLYHEVFDTLGSDESPS-------------
TH_0887|M.thermoresistible__bu LSFEDIPRVEQVHALLYHEVFDTLGSEESPS-------------
Mflv_4749|M.gilvum_PYR-GCK LEFQDIPRVEYVHSLLYHEVFDTLGSEESPS-------------
Mvan_1707|M.vanbaalenii_PYR-1 LEFQDIPRVEHVHSLLYHEVFDTLGSEESPS-------------
MAB_3625c|M.abscessus_ATCC_199 LEFHEIPRVEYVHSLLYHEVFDTLGSEEGDSRSTGRGRERPPAT
Rv3278c|M.tuberculosis_H37Rv LEFYNIPRLREVHALLYHEVFDTLGSDESPS-------------
Mb3306c|M.bovis_AF2122/97 LEFYNIPRLREVHALLYHKVFDTLGSDESPS-------------
MMAR_1258|M.marinum_M LEFYNIPRLREVHALLYHEVFDTLGSEEAPS-------------
MUL_2630|M.ulcerans_Agy99 LEFYNIPRLREVHALLYHEVFDTLGSEEAPS-------------
MLBr_00733|M.leprae_Br4923 VEFYNIPRLRQMYALLYHEVFDTLGSEESPS-------------
MAV_4246|M.avium_104 LEFHDIPRLREVHALLYHEVFDTLGSEESPS-------------
:.* :***:. :::****:*******:*. *