For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVAARLPFGWSADSGVTADIIEAAMELAIDTARHATAPFGAALLDVTTLRAFSGGNTYFESGDRFAHAET NVLRAAMSTLPELSNHVLISTAEPCPMCAAASVLSGVRAIIFGTSIETLIQCGWFQIRISASDVVAASTR PTRPSVYSGFLSHKTDLLYRNSENRRAMNPWTDPSH
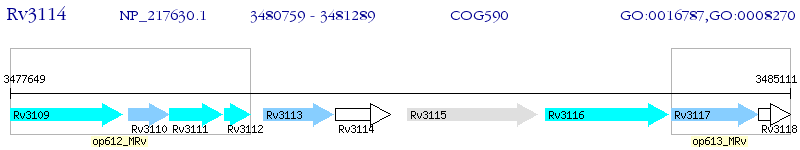
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3114 | - | - | 100% (176) | hypothetical protein Rv3114 |
| M. tuberculosis H37Rv | Rv3752c | - | 5e-08 | 32.37% (139) | cytidine/deoxycytidylate deaminase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3141 | - | 4e-98 | 99.43% (176) | hypothetical protein Mb3141 |
| M. gilvum PYR-GCK | Mflv_1214 | - | 1e-08 | 33.82% (136) | CMP/dCMP deaminase, zinc-binding |
| M. leprae Br4923 | MLBr_02474 | - | 9e-10 | 33.81% (139) | putative cytidine/deoxycytidylate deaminase |
| M. abscessus ATCC 19977 | MAB_0268 | - | 7e-09 | 33.09% (139) | cytidine/deoxycytidylate deaminase family protein |
| M. marinum M | MMAR_5295 | - | 3e-09 | 33.81% (139) | cytidine/deoxycytidylate deaminase |
| M. avium 104 | MAV_0324 | - | 2e-08 | 32.37% (139) | cytidine and deoxycytidylate deaminase family protein |
| M. smegmatis MC2 155 | MSMEG_6327 | - | 4e-07 | 33.33% (135) | cytidine and deoxycytidylate deaminase family protein |
| M. thermoresistible (build 8) | TH_1653 | - | 8e-08 | 33.58% (137) | POSSIBLE CYTIDINE/DEOXYCYTIDYLATE DEAMINASE |
| M. ulcerans Agy99 | MUL_4370 | - | 2e-09 | 33.81% (139) | cytidine/deoxycytidylate deaminase |
| M. vanbaalenii PYR-1 | Mvan_5594 | - | 4e-08 | 33.82% (136) | CMP/dCMP deaminase, zinc-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02474|M.leprae_Br4923 ---MPSAAGRRHQTRSVTTDEDLIRAALTVATTAG-SRDVPIGAVVLGAD
MAV_0324|M.avium_104 ----------------MISDEDLIRSALAVAATAG-PRDVPIGAVVVGAD
MMAR_5295|M.marinum_M ----------------MIADEDLVRAALAAAATAG-PRDVPIGAVVISAD
MUL_4370|M.ulcerans_Agy99 ----------------MIADEDLVRAALAAAATAG-PRDVPIGAVMISAD
MSMEG_6327|M.smegmatis_MC2_155 -----------------MTPESQIRAALEAARTAG-PRDVPIGAVVYAAD
Mflv_1214|M.gilvum_PYR-GCK -----------------MTDEDLIRAALDAARGAG-PRDVPIGAVVFDAE
Mvan_5594|M.vanbaalenii_PYR-1 MNRQFSSAGEPAIQSKQVTDEELIRAALDAARLAG-PRDVPIGAVVFAPD
TH_1653|M.thermoresistible__bu -------------MIGSVTDEDMIRAALTAAREAG-PRDVPIGAVVFGPD
MAB_0268|M.abscessus_ATCC_1997 --------------MTFPDDEALIRAALVAAREAG-SQDVPIGAVIYAAD
Rv3114|M.tuberculosis_H37Rv ---MVAARLPFGWSADSGVTADIIEAAMELAIDTARHATAPFGAALLDVT
Mb3141|M.bovis_AF2122/97 ---MVAARLPFGWPADSGVTADIIEAAMELAIDTARHATAPFGAALLDVT
:.:*: * :. .*:**.:
MLBr_02474|M.leprae_Br4923 GNELARAVNAREAIGDPTAHAEILAMRAAAGTLGNGWRLEGTTLAVTVEP
MAV_0324|M.avium_104 GTELARAVNAREALGDPTAHAEILALRAAAAALGDGWRLEGATLAVTVEP
MMAR_5295|M.marinum_M GTELARAVNAREELGDPTAHAEILALRAAARVLGDGWRLEGATLAVTVEP
MUL_4370|M.ulcerans_Agy99 GTELARAVNAREELGDPTAHAEILALRAAARVLGDGWRLEGATLAVTVEP
MSMEG_6327|M.smegmatis_MC2_155 GTELARAVNVREAYGDPAGHAEIVAMRAAAKVLGDGWRLEGATLAVTVEP
Mflv_1214|M.gilvum_PYR-GCK GTELARAANAREELGDPTAHAEILALRAAALVHGDGWRLEGATLAVTVEP
Mvan_5594|M.vanbaalenii_PYR-1 GRELARAANAREEFGDPTAHAEILALRAAARELGDGWRLEGATLAVTVEP
TH_1653|M.thermoresistible__bu GGELARAANAREQLGDPTAHAEVLALRAAARRLGDGWRLEGATLAVTVEP
MAB_0268|M.abscessus_ATCC_1997 GTELARAANARERLGDPTAHAEVLALRAAAAKHGDGWRLEGATLAVTVEP
Rv3114|M.tuberculosis_H37Rv TLRAFSGGNTYFESGDRFAHAETNVLRAAMSTLP---ELSNHVLISTAEP
Mb3141|M.bovis_AF2122/97 TLRAFSGGNTYFESGDRFAHAETNVLRAAMSTLP---ELSNHVLISTAEP
. . *. ** .*** .:*** .*.. .* *.**
MLBr_02474|M.leprae_Br4923 CTMCAGALVLARIERLVFGAWQPKTGAVG------SLWDVVRDHRLNHRP
MAV_0324|M.avium_104 CTMCAGALVLARIARLVFGAWEPKTGAVG------SLWDVVRDRRLNHRP
MMAR_5295|M.marinum_M CTMCAGALVLARVARLVFGAWEPKTGAVG------SLWDVVRDRRLNHRP
MUL_4370|M.ulcerans_Agy99 CTMCAGALVLARVARLVFGAWEPKTGAVG------SLWDVVRDRRLNHRP
MSMEG_6327|M.smegmatis_MC2_155 CTMCAGALVLARVSRLVFGAWEPKTGAVG------SLWDVVRDRRLNHRP
Mflv_1214|M.gilvum_PYR-GCK CTMCAGALVMARVARVVFGAWEPKTGAVG------SLWDVVRDRRLTHRP
Mvan_5594|M.vanbaalenii_PYR-1 CTMCAGALVMARVARVVFGAWEPKTGAVG------SLWDVVRDRRLTHRP
TH_1653|M.thermoresistible__bu CTMCAGALVLARVARLVFGAWEPKTGAVG------SLWDVVRDRRLNHRP
MAB_0268|M.abscessus_ATCC_1997 CTMCAGALVLARVGRVVFGAWEPKTGAVG------SLWDVVRDRRQAHRP
Rv3114|M.tuberculosis_H37Rv CPMCAAASVLSGVRAIIFGTSIETLIQCGWFQIRISASDVVAASTRPTRP
Mb3141|M.bovis_AF2122/97 CPMCAAASVLSGVRAIIFGTSIETLIQCGWFQIRISASDVVAASTRPTRP
*.***.* *:: : ::**: . * * *** **
MLBr_02474|M.leprae_Br4923 AVRGGVLAQECTAPLEAFFAHQRLSKGPPVR-
MAV_0324|M.avium_104 EVRGGVLERECAALLEGFFARQRLG-------
MMAR_5295|M.marinum_M QVRGGVLAQECAAPLEEFFGRQRLG-------
MUL_4370|M.ulcerans_Agy99 QVRGGVLAQECAAPLEEFFGRQRLG-------
MSMEG_6327|M.smegmatis_MC2_155 EVIGGVLADECAALLEEFFARQR---------
Mflv_1214|M.gilvum_PYR-GCK QVRGGVLEAECAAPLEEFFARQR---------
Mvan_5594|M.vanbaalenii_PYR-1 QVRGGVLEAECAAPLEEFFAGQR---------
TH_1653|M.thermoresistible__bu EVRGGVLAAECAAPLEEFFARQRDPDSPRGLG
MAB_0268|M.abscessus_ATCC_1997 EVRGGVLEHECAALLERFFAAKR---------
Rv3114|M.tuberculosis_H37Rv SVYSGFLSHKTDLLYRNSENRRAMNPWTDPSH
Mb3141|M.bovis_AF2122/97 SVYSGFLSHKTDLLYRNSENRRAMNPWTDPSH
* .*.* : . :