For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KDRVGFLRGRARPASTLITRFIADHQGHREGPDGLRWGVESICTQLTELGVPIAPSTYYDHINREPSRRE LRDGELKEHISRVHAANYGVYGARKVWLTLNREGIEVARCTVERLMTKLGLSGTTRGKARRTTIADPATA RPADLVQRRFGPPAPNRLWVADLTYVSTWAGFAYVAFVTDAYARRILGWRVASTMATSMVLDAIEQAIWT RQQEGVLDLKDVIHHTDRGSQYTSIRFSERLAEAGIQPSVGAVGSSYDNALAETINGLYKTELIKPGKPW RSIEDVELATARWVDWFNHRRLYQYCGDVPPVELEAAYYAQRQRPAAG
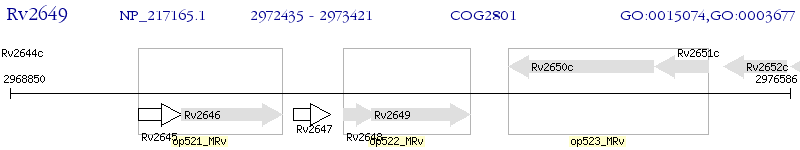
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2649 | - | - | 100% (328) | PROBABLE TRANSPOSASE FOR INSERTION SEQUENCE ELEMENT IS6110 |
| M. tuberculosis H37Rv | Rv3475 | - | 0.0 | 100.00% (328) | transposase IS6110 |
| M. tuberculosis H37Rv | Rv2479c | - | 0.0 | 100.00% (328) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2838c | - | 1e-163 | 100.00% (278) | transposase |
| M. gilvum PYR-GCK | Mflv_3221 | - | 2e-91 | 51.38% (325) | integrase catalytic subunit |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2086c | - | 4e-29 | 55.96% (109) | putative transposase-like protein IS1601-D |
| M. marinum M | MMAR_2548 | - | 4e-81 | 51.06% (284) | transposase, ISMyma03_aa2 |
| M. avium 104 | MAV_5093 | - | 5e-83 | 53.26% (291) | transposase |
| M. smegmatis MC2 155 | MSMEG_1721 | - | 9e-84 | 50.00% (310) | IS1137, transposase orfB |
| M. thermoresistible (build 8) | TH_3317 | - | 2e-76 | 50.67% (298) | PUTATIVE Integrase, catalytic region |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5973 | - | 1e-57 | 54.03% (211) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2649|M.tuberculosis_H37Rv ----------------------------------------KDRVGFLRGR
Mb2838c|M.bovis_AF2122/97 --------------------------------------------------
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 --------------------------------------------------
TH_3317|M.thermoresistible__bu --------------------------------------------------
Mvan_5973|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3221|M.gilvum_PYR-GCK MGPPGRDRRRRSGWADQRGIRGPAQAAPGERRTQAGQRDFEGGVGFLRCR
MSMEG_1721|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2548|M.marinum_M --------------------------------------------------
Rv2649|M.tuberculosis_H37Rv ARPASTLITRFIADHQGHREGPDGLRWGVESICTQLT-ELGVPIAPSTYY
Mb2838c|M.bovis_AF2122/97 -----------------------------------------MPIAPSTYY
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 --------MAFIDANR--------DEFGVEPICTVLRSAGLQVAPST-YY
TH_3317|M.thermoresistible__bu -------MIAFIDAHR--------DQFGVELICRVLRAAIAGFLTSRGYR
Mvan_5973|M.vanbaalenii_PYR-1 -------------------------------------------MAPSTYY
Mflv_3221|M.gilvum_PYR-GCK ARPAVSVVVEFISAHQHMRVGADGLKWGVESMC-AVLSEFGVVIAPSTYY
MSMEG_1721|M.smegmatis_MC2_155 --------MEFISAHQHMRVGVDGLKWGVESMC-AVLSEYGVTIAPSTYY
MMAR_2548|M.marinum_M -----------------------------------MLTEHGIKISPSTYY
Rv2649|M.tuberculosis_H37Rv DHINRE-PSRRELRDGELKEHISRVHAAN--YGVYGARKVWLTLNREGIE
Mb2838c|M.bovis_AF2122/97 DHINRE-PSRRELRDGELKEHISRVHAAN--YGVYGARKVWLTLNREGIE
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 ANKTRT-PSARAQRDAVMRPVITQLWEDN--YRVYGARKLWKAARRAGHD
TH_3317|M.thermoresistible__bu AAKARP-PSERAMRDELLIAELRTVHEQN--FSVYGVKKMHHAMKRRGWR
Mvan_5973|M.vanbaalenii_PYR-1 DVKSRC-PSARARRDAELGPALRVIWEDN--YRVYGAVKLWKAARRGGHD
Mflv_3221|M.gilvum_PYR-GCK AHRARRGPSKADWTDAQVIDAIWRLRRSNKLFAVLGARKTWIVLRTNGLD
MSMEG_1721|M.smegmatis_MC2_155 AHRARQGPSKADWADAQVIDAIWQLRQSQSLYRVLGARKTWIVLRTNGID
MMAR_2548|M.marinum_M EWMKRT-PSKQQLRDEAVTASIKEFRERNPLNKTLGSRKVWIKLRGDGMD
Rv2649|M.tuberculosis_H37Rv VARCTVERLMTKLGLSGTTRGKARRTTIADPATARPADLVQRRFGPPAPN
Mb2838c|M.bovis_AF2122/97 VARCTVERLMTKLGLSGTTRGKARRTTIADPATARPADLVQRRFGPPAPN
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 IGRDQIARLMREAGIAGVRRGKRVRTTKPGPGAPRHPDLVGRDFTASAPN
TH_3317|M.thermoresistible__bu VGREQTRRLMRKAGLRGVQRGKPVFTTISDPAHCRPADLVNRQFSAEAPD
Mvan_5973|M.vanbaalenii_PYR-1 VGRDQVARLMRSLGIEGVRRGKRVTTTRADPAAPRHPDLVKRQFTATAPN
Mflv_3221|M.gilvum_PYR-GCK VSRCVVERVMREMGWRGACKRRRVRTTIADPAATRAPDRVARHFVAGAPD
MSMEG_1721|M.smegmatis_MC2_155 VSRCVVERVMREMGWRGACKRRRVRTTVADPAATRAPDRVRRNFVAGAPD
MMAR_2548|M.marinum_M VARCTVERLMQANRWQGARYGGAPKTTIQDEKAQRATDLVDRVFAAPAPN
Rv2649|M.tuberculosis_H37Rv RLWVADLTYVSTWAGFAYVAFVTDAYARRILGWRVASTMATS-MVLDAIE
Mb2838c|M.bovis_AF2122/97 RLWVADLTYVSTWAGFAYVAFVTDAYARRILGWRVASTMATS-MVLDAIE
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 QLWVTDLTFVPTWAGVAYVCFLTDAFSRMIIGWRVASHMRTT-VVLDTIE
TH_3317|M.thermoresistible__bu RLWVADITFVRTWQGFCYTAFVTDACTKAIKGWAVAASMRTEDLPLEAFN
Mvan_5973|M.vanbaalenii_PYR-1 QLWVTDLTFVPTWVGVAYVCFIVDAFSRLIVGWRAASNMRTT-MVLDALE
Mflv_3221|M.gilvum_PYR-GCK RLWVADFTYCRTRAGWAYTAFVTDVYARKIVGWKVATEMTQK-LVTDAIN
MSMEG_1721|M.smegmatis_MC2_155 RLWVADFTYCRTRAGWAYTAFVTDVYARKIVGWKVATEMTQK-LVTDAID
MMAR_2548|M.marinum_M RLWVADFTYCPTWAGMVYVAFVIDAFARRVIGWRAATSMKTA-LVIDALE
Rv2649|M.tuberculosis_H37Rv QAIWTRQQEGVLDLKDVIHHTDRGSQYTSIRFSERLAEAGIQPSVGAVGS
Mb2838c|M.bovis_AF2122/97 QAIWTRQQEGVLDLKDVIHHTDRGSQYTSIRFSERLAEAGIQPSVGAVGS
MAB_2086c|M.abscessus_ATCC_199 -------------MPGLTCHSDAGSQFTSIRYGERLAEIGAVPSIGTIGD
MAV_5093|M.avium_104 MARWSRGK----MLAGLRCHSDAGSQFTSIRYSERLAEIGAVPSIGTVGD
TH_3317|M.thermoresistible__bu HAVWHSDS----DLSELIHHSDRGSQYLSLTYTDRLAELGIAPSVGSRGD
Mvan_5973|M.vanbaalenii_PYR-1 MARWSRGN----TLSGLRCHSDAGSQFTSIRYGERLAELGAVPSIGSVGD
Mflv_3221|M.gilvum_PYR-GCK HAIDTRKRSGATFLDDLIHHSDAGSQYTAVAFTEHLAAEGILPSVGSVGD
MSMEG_1721|M.smegmatis_MC2_155 HAIDTRKRSGAASLDSLIHHSDAGSQYTAVAFTERLAAEGILPSVGSVGD
MMAR_2548|M.marinum_M HAFFTRAQEGFTDLSGLVAHSDAGSQYTSIAFTTRLIDAGIDASVGSVAD
: : *:* ***: :: : :* * .*:*: ..
Rv2649|M.tuberculosis_H37Rv SYDNALAETINGLYKTELIK---PGKPWRSIEDVELATARWVDWFNHRRL
Mb2838c|M.bovis_AF2122/97 SYDNALAETINGLYKTELIK---PGKPWRSIEDVELATARWVDWFNHRRL
MAB_2086c|M.abscessus_ATCC_199 SYDNAL-ETVNGYYKAELIYGPVRTRPWKTVEDVELATLSWVYWHNTSRL
MAV_5093|M.avium_104 SFDNALAETVNGYYKAELIYGPARPGPWKTVEDVELATLSWVHWHNTSRL
TH_3317|M.thermoresistible__bu SYDNALAEAVNAAYKTELIN---RGKPWRSVDDVELATAGWVAWYNQERL
Mvan_5973|M.vanbaalenii_PYR-1 SFDNALAERAC------LID----------------CVSGWA--------
Mflv_3221|M.gilvum_PYR-GCK SFDNALAESVNSSYKTELID---HQPPYPGAAELSLATAEWVAFYNRQRP
MSMEG_1721|M.smegmatis_MC2_155 SFDNALAESVNSSYKTELID---RQPLYPGATELALGTAEWVAFYNRQRP
MMAR_2548|M.marinum_M AYDNALAETTVGSFKNELIR---RQGPWRDVDHVEIGTLNWVDWFNNERP
::**** * ** . *.
Rv2649|M.tuberculosis_H37Rv YQYCGDVPPVELEAAYYAQRQRPAAG-------
Mb2838c|M.bovis_AF2122/97 YQYCGDVPPVELEAAYYAQRQRPAAG-------
MAB_2086c|M.abscessus_ATCC_199 HSYLNDIPPTEFEAAFY-DAHGTDQPLAGIQ--
MAV_5093|M.avium_104 HGYLNDVPPAEFEAAFY-ATKRTDHPLVEIQ--
TH_3317|M.thermoresistible__bu HEALGYVPPAEYEAALTGTSHPASQPTPALATN
Mvan_5973|M.vanbaalenii_PYR-1 ---------------------------------
Mflv_3221|M.gilvum_PYR-GCK NGYCQDLTPDRAEALYYHRQRHPHTEEALR---
MSMEG_1721|M.smegmatis_MC2_155 NGYCQDLTPDRAEALYYHRQRHPHAEEALR---
MMAR_2548|M.marinum_M HEYLADLTPTQAEALHYRHKSSLAAAG------