For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SASLLVRTACGGRAVAQRLRTVLWPITQTSVVAGLAWYLTHDVFNHPQAFFAPISAVVCMSATNVLRARR AQQMIVGVALGIVLGAGVHALLGSGPIAMGVVVFIALSVAVLCARGLVAQGLMFINQAAVSAVLVLVFAS NGSVVFERLFDALVGGGLAIVFSILLFPPDPVVMLCSARADVLAAVRDILAELVNTVSDPTSAPPDWPMA AADRLHQQLNGLIEVRANAAMVARRAPRRWGVRSTVRDLDQQAVYLALLVSSVLHLARTIAGPGGDKLPT PVHAVLTDLAAGTGLADADPTAANEHAAAARATASTLQSAACGSNEVVRADIVQACVTDLQRVIERPGPS GMSA*
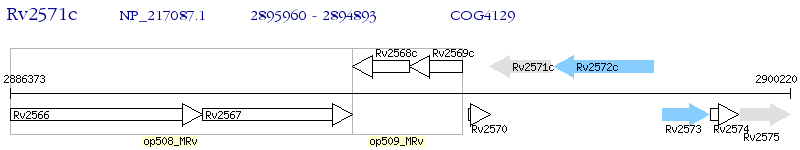
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2571c | - | - | 100% (355) | PROBABLE TRANSMEMBRANE ALANINE AND VALINE AND LEUCINE RICH PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2601c | - | 0.0 | 100.00% (355) | transmembrane alanine and valine and leucine rich protein |
| M. gilvum PYR-GCK | Mflv_0746 | - | 3e-08 | 24.79% (242) | membrane protein-like protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2160 | - | 1e-114 | 62.14% (346) | transmembrane alanine and valine and leucine rich protein |
| M. avium 104 | MAV_3441 | - | 5e-88 | 51.30% (347) | hypothetical protein MAV_3441 |
| M. smegmatis MC2 155 | MSMEG_0090 | - | 1e-06 | 27.61% (163) | hypothetical protein MSMEG_0090 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK -MLS---LRQRVADRVAQRDPDRDALRRAVRAAVVVPAAGAVSFAVAGGS
MSMEG_0090|M.smegmatis_MC2_155 MLLTPAKLWRRAAERLRERDPEYDALRRAARAGVVLPIAAAIGFAVGGGS
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK TQTPLFTIFGAVALLVFSDFPGNRQNRAVAYGGLGLNGFLLITLGTLVAP
MSMEG_0090|M.smegmatis_MC2_155 -QTPLFSIFGAVSLLITADFPGNRPARALAFAGLAFNGAVLIVLGTLIAP
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK YPWPAVAAMFVIGVAVTFSGVLSETLAAGQRATLLTFVLPACTPPGPVAE
MSMEG_0090|M.smegmatis_MC2_155 HVWLSVVSMFVIGVVVSFSGVLSEVVAAGQRATLLLFVLPLCTPVGPIAD
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK RLLGWTIALAVCVPAALFLLPPRHHGQLRRRAARACTALADRLDGTVTPD
MSMEG_0090|M.smegmatis_MC2_155 RLLGWVIALVVCVPAALFLFPPRHHDELRRTAARVCRVLAYRLEGSASAR
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK DVQRAMDALRKTFLGADFRPVGLTAGSRALVRVVDDLEWVADRTGRGTGV
MSMEG_0090|M.smegmatis_MC2_155 DVNRAMNELYESFLGADYRPVALTAGSRALVRVVDDLGWISDRVTDSTGE
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK TLRDK-DAVVRVLRASAAVLSISRPADREMARAELEGALLALRTLARGRW
MSMEG_0090|M.smegmatis_MC2_155 ILGEMRDPVVRVLRDSAALLRIHDPARRAERSADLRAALEELRTVSQGTY
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
Mflv_0746|M.gilvum_PYR-GCK REDIDEILGAADDEQAVAQGRDLLRRRTIAATVSATGRVIAAAAAADARP
MSMEG_0090|M.smegmatis_MC2_155 REGIAAILGTPDDADAVEVGKALLVRRTIAATVAVTGRVIRNAALADARP
Rv2571c|M.tuberculosis_H37Rv -------------MSASLLVRTACGGRAVAQRLRTVLWPITQTSVVAGLA
Mb2601c|M.bovis_AF2122/97 -------------MSASLLVRTACGGRAVAQRLRTVLWPITQTSVVAGLA
MMAR_2160|M.marinum_M -------------MEISLLTRAADGSLAAVRQLRTALWPITQASVTAGLA
MAV_3441|M.avium_104 ----------MRGSAVPSLVRTAA--RAGGKRLGAVWFNLLQTSLAAGLS
Mflv_0746|M.gilvum_PYR-GCK VWARALGLRLPPTGASDRLLPETVAAAKITTGFVENRSVAVRNSLRTGLG
MSMEG_0090|M.smegmatis_MC2_155 VWARVLGRRLPETGSADWIMPETTAVAEIAKGFLSTRAVVLRNSLRTGLG
: : : : *: :**.
Rv2571c|M.tuberculosis_H37Rv WYLTH---DVFNHPQAFFAPISAVVCMSATNVLRARRAQQMIVGVALGIV
Mb2601c|M.bovis_AF2122/97 WYLTH---DVFNHPQAFFAPISAVVCMSATNVLRARRAQQMIVGVALGIV
MMAR_2160|M.marinum_M WFLTH---DVLDHPQPFFAPISAAVCMSATNVLRARRAVQMVGGVALGIV
MAV_3441|M.avium_104 WYLAH---DVLDHPQPFFAPIAAAVSLSTSNVLRAQRAVQMMIGVTLGIG
Mflv_0746|M.gilvum_PYR-GCK LALAVTVTHVFPVEHGFWVVLGALSVLRSSALTTGTRVWRAVVGTGIGFL
MSMEG_0090|M.smegmatis_MC2_155 LALAVAVTHVFPVEHGFWVVLGAMSVLRSSALTTGTRVVRAVAGTALGFV
*: .*: : *:. :.* : :: : . *. : : *. :*:
Rv2571c|M.tuberculosis_H37Rv LGAGVHALLGSGPIAMGVVVFIALSVAVLCARGLVAQGLMFINQAAVSAV
Mb2601c|M.bovis_AF2122/97 LGAGVHALLGSGPIAMGVVVFIALSVAVLCARGLVAQGLMFINQAAVSAV
MMAR_2160|M.marinum_M VGAGVQAVLGTGPTAMGVAVFAALSVAVLAVRGFVAQGLMFVNQTAVSAV
MAV_3441|M.avium_104 LGTVVQGMLGPGALPIAVAALVALGAAVFIGGGFIGHGMMFANQTVVSAL
Mflv_0746|M.gilvum_PYR-GCK LGALLISLVGVDPVVMWILMPLAVFGSAYVPEIASFIAAQAAFTMMVLIS
MSMEG_0090|M.smegmatis_MC2_155 IGAVLIELVGVQPVVLWILLPIVAFGSAYVPEVASFIAGQAAFTMMVLIN
:*: : ::* . : : . :. . *
Rv2571c|M.tuberculosis_H37Rv LVLVFASNGSVVFERLFDALVGGGLAIVFSILLFPPDPVVMLCSARADVL
Mb2601c|M.bovis_AF2122/97 LVLVFASNGSVVFERLFDALVGGGLAIVFSILLFPPDPVVMLCSARADVL
MMAR_2160|M.marinum_M LVLVFAPTGDVVAERLFDALIGGGLALVVAMLLFPADPVRILRDARTGAL
MAV_3441|M.avium_104 LVLALYRGG-AGPERIFDALIGGAVAIVVAVLLFPADPRTVLGAARAGVL
Mflv_0746|M.gilvum_PYR-GCK FNLIAPTGWEVGLIRVEDVFVGALVGVVVSVLLWPRGATASVTRAIDSAR
MSMEG_0090|M.smegmatis_MC2_155 FNLIAPTGWQVGLIRIEDVVVGALVGIVVSILLWPRGARASVTKAIDAAR
: * . *: *..:*. :.:*.::**:* .. : * .
Rv2571c|M.tuberculosis_H37Rv AAVRDILAELVNTVSDPTSAPPDWPMAAADRLHQQLNGLIEVRANAAMVA
Mb2601c|M.bovis_AF2122/97 AAVRDILAELVNTVSDPTSAPPDWPMAAADRLHQQLNGLIEVRANAAMVA
MMAR_2160|M.marinum_M AALHDTLVEVIKFIDNPGGAAPDWLLATFDRLHNQLGGLIEARATAVLVS
MAV_3441|M.avium_104 AVLHDVLSRAADVSSGRRAAPPDWPLSAVDRVHEQLSGLLEARTTARHVV
Mflv_0746|M.gilvum_PYR-GCK FVAARYLRAAVLRIT--RGAYEERTNEVVTLGHAALDASRVVDDAVRHYL
MSMEG_0090|M.smegmatis_MC2_155 AAGAELLEAAVLRVT--RGASEEATDRVIALSHSALEASRTLDDTVRHYL
. * .* : . * * . .
Rv2571c|M.tuberculosis_H37Rv RRAPRRWGVRSTVRDLDQQAVYLALLVSSVLHLARTIAGPGGDKLPTPVH
Mb2601c|M.bovis_AF2122/97 RRAPRRWGVRSTVRDLDQQAVYLALLVSSVLHLARTIAGPGGDKLPTPVH
MMAR_2160|M.marinum_M RRAPRRWAARDAIADIERQSARLGMLVSGVLNLVRAITRLPEQQVPRPVR
MAV_3441|M.avium_104 AIAPRRWGLRDAVRAADHQAVHVALLAGSVLQLARAVAPGPGDRQGQPVS
Mflv_0746|M.gilvum_PYR-GCK SESSGETDFRAPVVRSSNRATRLRGAADLIADIP-TPPPLGTYPKVRSVL
MSMEG_0090|M.smegmatis_MC2_155 SESGGPSDQRAPIVRSATRAIRVRAAAELIADV--VPPPLEAYQETRKTI
: * .: :: : . : .: . . .
Rv2571c|M.tuberculosis_H37Rv AVLTDLAAGTGLADADPTAANEHAAAARATASTLQSAACGSNE--VVRAD
Mb2601c|M.bovis_AF2122/97 AVLTDLAAGTGLADADPTAANEHAAAARATASTLQSAACGSNE--VVRAD
MMAR_2160|M.marinum_M IALAELAAALAVADDEPAAAIAHAAAARDRAQELLSQARDRTE--VVLAD
MAV_3441|M.avium_104 TVLLVLAAATALADRDPAGACVYLGSARRHAARLRSGDGGDREPHVALAD
Mflv_0746|M.gilvum_PYR-GCK EAHADAICARVSGTSEPVGAWAPISEDFVRALRSEAPEDTLGVAAALPLL
MSMEG_0090|M.smegmatis_MC2_155 EAHARIICARLTG-EDTTSVFVPIGETFVLALRAEATGGDLAVSAALPLV
. .. . :.... . * : .
Rv2571c|M.tuberculosis_H37Rv IVQACVTDLQRVIER-PGPSGMSA
Mb2601c|M.bovis_AF2122/97 IVQACVTDLQRVIER-PGPSGMSA
MMAR_2160|M.marinum_M IVDACGVDLQRVIDV-PGGDH---
MAV_3441|M.avium_104 AVGACVDDLQRVIDLRPG------
Mflv_0746|M.gilvum_PYR-GCK TAAAALGELELIYPWDTGPS----
MSMEG_0090|M.smegmatis_MC2_155 TVAAHIGELELLYPQPVESVH---
. * :*: :