For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RGSAVPSLVRTAARAGGKRLGAVWFNLLQTSLAAGLSWYLAHDVLDHPQPFFAPIAAAVSLSTSNVLRAQ RAVQMMIGVTLGIGLGTVVQGMLGPGALPIAVAALVALGAAVFIGGGFIGHGMMFANQTVVSALLVLALY RGGAGPERIFDALIGGAVAIVVAVLLFPADPRTVLGAARAGVLAVLHDVLSRAADVSSGRRAAPPDWPLS AVDRVHEQLSGLLEARTTARHVVAIAPRRWGLRDAVRAADHQAVHVALLAGSVLQLARAVAPGPGDRQGQ PVSTVLLVLAAATALADRDPAGACVYLGSARRHAARLRSGDGGDREPHVALADAVGACVDDLQRVIDLRP G*
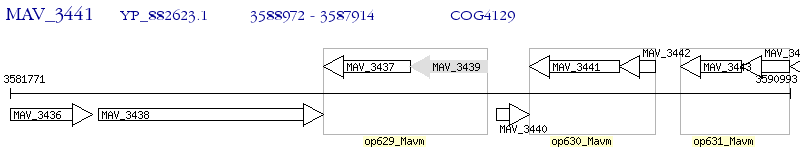
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3441 | - | - | 100% (352) | hypothetical protein MAV_3441 |
| M. avium 104 | MAV_4931 | - | 2e-70 | 45.07% (335) | hypothetical protein MAV_4931 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2601c | - | 4e-88 | 51.30% (347) | transmembrane alanine and valine and leucine rich protein |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2571c | - | 4e-88 | 51.30% (347) | transmembrane alanine and valine and leucine rich protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2160 | - | 1e-79 | 49.70% (338) | transmembrane alanine and valine and leucine rich protein |
| M. smegmatis MC2 155 | MSMEG_0090 | - | 3e-06 | 25.24% (313) | hypothetical protein MSMEG_0090 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3048 | - | 6e-06 | 30.66% (137) | protein of unknown function DUF893, YccS/YhfK |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
MSMEG_0090|M.smegmatis_MC2_155 MLLTPAKLWRRAAERLRERDPEYDALRRAARAGVVLPIAAAIGFAVGGGS
Mvan_3048|M.vanbaalenii_PYR-1 --------------MIKGLVQRSAMPDPAAVLRSLACVAAVAVVAVQWGP
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
MSMEG_0090|M.smegmatis_MC2_155 QTPLFSIFGAVSLLITADFPGNRPARALAFAGLAFNGAVLIVLGTLIA--
Mvan_3048|M.vanbaalenii_PYR-1 QGSATAVVGAAAVAGATALQDSPRGRVTVVLLVTAVAGLAVLLGALTSGF
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
MSMEG_0090|M.smegmatis_MC2_155 PHVWLSVVSMFVIGVVVSFSGVLSEVVAAGQRATLLLFVLPLCTPVGPIA
Mvan_3048|M.vanbaalenii_PYR-1 SPLFVAVATLWCFGASMLWALGANPGLIGAASAALLVSAPPVTPTPTSVV
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
MSMEG_0090|M.smegmatis_MC2_155 DRLLGWVIALVVCVPAALFLFPPRHHDELRRTAARVCRVLAYRLEGSASA
Mvan_3048|M.vanbaalenii_PYR-1 VTWLLAVAGGLLQVWVISLWPPLRWRNQRKALVAAYR-------------
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
MSMEG_0090|M.smegmatis_MC2_155 RDVNRAMNELYESFLGADYRPVALTAGSRALVRVVDDLGWISDRVTDSTG
Mvan_3048|M.vanbaalenii_PYR-1 -------------SLAADARKISDGTVGGGPAADPEPLAALRTAFTAADG
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
MSMEG_0090|M.smegmatis_MC2_155 EILGEMRDPVVRVLRDSAALLRIHDPARRAERSADLRAALEELRTVSQGT
Mvan_3048|M.vanbaalenii_PYR-1 PSRRRPATYRGWYALPERIADTLTDVARLPARSDALALALAEAADTLAAV
Mb2601c|M.bovis_AF2122/97 --------------------------------------------------
Rv2571c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2160|M.marinum_M --------------------------------------------------
MAV_3441|M.avium_104 --------------------------------------------------
MSMEG_0090|M.smegmatis_MC2_155 YREGIAAILGTPDDADAVEVGKALLVRRTIAATVAVTGRVIRNAALADAR
Mvan_3048|M.vanbaalenii_PYR-1 ADTSRTGRVG----ADVAIGRFDSVAATVDGSESVVVQKLSTQLHEAVAM
Mb2601c|M.bovis_AF2122/97 --------------MSASLLVRTACGGRAVAQRLRTVLWPITQTSVVAGL
Rv2571c|M.tuberculosis_H37Rv --------------MSASLLVRTACGGRAVAQRLRTVLWPITQTSVVAGL
MMAR_2160|M.marinum_M --------------MEISLLTRAADGSLAAVRQLRTALWPITQASVTAGL
MAV_3441|M.avium_104 -----------MRGSAVPSLVRTAA--RAGGKRLGAVWFNLLQTSLAAGL
MSMEG_0090|M.smegmatis_MC2_155 PVWARVLGRRLPETGSADWIMPETTAVAEIAKGFLSTRAVVLRNSLRTGL
Mvan_3048|M.vanbaalenii_PYR-1 RLGDFVPSTPDAVRLRRPELRTSVRSAVDVMRSHLDRHSPVLRHAVRLAA
: . : : : :: .
Mb2601c|M.bovis_AF2122/97 AWYLT---HDVFNHPQAFFAPISAVVCMSATNVLRARRAQQMIVGVALGI
Rv2571c|M.tuberculosis_H37Rv AWYLT---HDVFNHPQAFFAPISAVVCMSATNVLRARRAQQMIVGVALGI
MMAR_2160|M.marinum_M AWFLT---HDVLDHPQPFFAPISAAVCMSATNVLRARRAVQMVGGVALGI
MAV_3441|M.avium_104 SWYLA---HDVLDHPQPFFAPIAAAVSLSTSNVLRAQRAVQMMIGVTLGI
MSMEG_0090|M.smegmatis_MC2_155 GLALAVAVTHVFPVEHGFWVVLGAMSVLRSSALTTGTRVVRAVAGTALGF
Mvan_3048|M.vanbaalenii_PYR-1 AVAVGCVVDRYAEGAYGFWIPLTVLMVLRPETAHTYTRCVGRVAGTLVGI
. : *: : . : . * : *. :*:
Mb2601c|M.bovis_AF2122/97 VLGAGVHALLGSGPIAMGVVVFIALSVAVLCARGLVAQGLMFINQAAVSA
Rv2571c|M.tuberculosis_H37Rv VLGAGVHALLGSGPIAMGVVVFIALSVAVLCARGLVAQGLMFINQAAVSA
MMAR_2160|M.marinum_M VVGAGVQAVLGTGPTAMGVAVFAALSVAVLAVRGFVAQGLMFVNQTAVSA
MAV_3441|M.avium_104 GLGTVVQGMLGPGALPIAVAALVALGAAVFIGGGFIGHGMMFANQTVVSA
MSMEG_0090|M.smegmatis_MC2_155 VIGAVLIELVGVQPVVLWILLPIVAFGSAYVPEVASFIAGQAAFTMMVLI
Mvan_3048|M.vanbaalenii_PYR-1 VTASAVLVILNPG-VAVSAVLALAAVGVACVLSGIGFIAMAAAVTAAAVF
.: : ::. : . . . .
Mb2601c|M.bovis_AF2122/97 VLVLVFASNGSVVFERLFDALVGGGLAIVFSILLFPPDPVVMLCSARADV
Rv2571c|M.tuberculosis_H37Rv VLVLVFASNGSVVFERLFDALVGGGLAIVFSILLFPPDPVVMLCSARADV
MMAR_2160|M.marinum_M VLVLVFAPTGDVVAERLFDALIGGGLALVVAMLLFPADPVRILRDARTGA
MAV_3441|M.avium_104 LLVLALYRGG-AGPERIFDALIGGAVAIVVAVLLFPADPRTVLGAARAGV
MSMEG_0090|M.smegmatis_MC2_155 NFNLIAPTGWQVGLIRIEDVVVGALVGIVVSILLWPRGARASVTKAIDAA
Mvan_3048|M.vanbaalenii_PYR-1 LVDIGRPDMPPAVADPVLAVLFGGALALIAHVVLP-DDALTRLSQRAGEL
. : . : .:.*. :.:: ::* .. :
Mb2601c|M.bovis_AF2122/97 LAAVRDILAELVNTVSDPTSAPPDWPMAAADRLHQQLNGLIEVRANAAMV
Rv2571c|M.tuberculosis_H37Rv LAAVRDILAELVNTVSDPTSAPPDWPMAAADRLHQQLNGLIEVRANAAMV
MMAR_2160|M.marinum_M LAALHDTLVEVIKFIDNPGGAAPDWLLATFDRLHNQLGGLIEARATAVLV
MAV_3441|M.avium_104 LAVLHDVLSRAADVSSGRRAAPPDWPLSAVDRVHEQLSGLLEARTTARHV
MSMEG_0090|M.smegmatis_MC2_155 RAAGAELLEAAVLRVTRGASEEATDRVIALS-----HSALEASRTLDDTV
Mvan_3048|M.vanbaalenii_PYR-1 LRTEIDYAATVIKAYVHELDDPAETMTSAWQR---------AFRARAAFE
. : . : . *:
Mb2601c|M.bovis_AF2122/97 ARRAPRRWGVRSTVRDLDQQAVYLALLVSSVLHLARTIAGPGGDKLPTPV
Rv2571c|M.tuberculosis_H37Rv ARRAPRRWGVRSTVRDLDQQAVYLALLVSSVLHLARTIAGPGGDKLPTPV
MMAR_2160|M.marinum_M SRRAPRRWAARDAIADIERQSARLGMLVSGVLNLVRAITRLPEQQVPRPV
MAV_3441|M.avium_104 VAIAPRRWGLRDAVRAADHQAVHVALLAGSVLQLARAVAPGPGDRQGQPV
MSMEG_0090|M.smegmatis_MC2_155 RHYLSESGGPSDQRAPIVRSATRAIRVRAAAELIADVVPPPLEAYQETRK
Mvan_3048|M.vanbaalenii_PYR-1 ATTGAMRMESRELRHWLRSYRTALNAVTSSCTTLEDQLPAQPAVTANREF
. . . : .. : :.
Mb2601c|M.bovis_AF2122/97 HAVLTDLAAGTGLADADPTAANEHAAAARATASTLQSAACGSNE--VVRA
Rv2571c|M.tuberculosis_H37Rv HAVLTDLAAGTGLADADPTAANEHAAAARATASTLQSAACGSNE--VVRA
MMAR_2160|M.marinum_M RIALAELAAALAVADDEPAAAIAHAAAARDRAQELLSQARDRTE--VVLA
MAV_3441|M.avium_104 STVLLVLAAATALADRDPAGACVYLGSARRHAARLRSGDGGDREPHVALA
MSMEG_0090|M.smegmatis_MC2_155 TIEAHARIICARLTGEDTTSVFVPIGETFVLALRAEATGGDLAVSAALPL
Mvan_3048|M.vanbaalenii_PYR-1 VLAVDEYVEALRGDPPTAASPWRVDCAELAAAEARLREAVRRHGPEEGPA
.:. *
Mb2601c|M.bovis_AF2122/97 DIVQACVTDLQRVIER-PGPSGMSA--
Rv2571c|M.tuberculosis_H37Rv DIVQACVTDLQRVIER-PGPSGMSA--
MMAR_2160|M.marinum_M DIVDACGVDLQRVIDV-PGGDH-----
MAV_3441|M.avium_104 DAVGACVDDLQRVIDLRPG--------
MSMEG_0090|M.smegmatis_MC2_155 VTVAAHIGELELLYPQPVESVH-----
Mvan_3048|M.vanbaalenii_PYR-1 RLLVAEVDTITRHVSAISVSPSPTSVR
: * :