For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIPNPLEELTLEQLRSQRTSMKWRAHPADVLPLWVAEMDVKLPPTVADALRRAIDDGDTGYPYGTEYAEA VREFACQRWQWHDLEVSRTAIVPDVMLGIVEVLRLITDRGDPVIVNSPVYAPFYAFVSHDGRRVIPAPLR GDGRIDLDALQEAFSSARASSGSSGNVAYLLCNPHNPTGSVHTADELRGIAERAQRFGVRVVSDEIHAPL IPSGARFTPYLSVPGAENAFALMSASKAWNLGGLKAALAIAGREAAADLARMPEEVGHGPSHLGVIAHTA AFRTGGNWLDALLRGLDHNRTLLGALVDEHLPGVQYRWPQGTYLAWLDCRELGFDDAASDEMTEGLAVVS DLSGPARWFLDHARVALSSGHVFGIGGAGHVRINFATSRAILIEAVSRMSRSLLERR
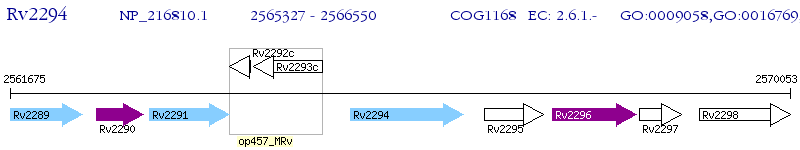
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2294 | - | - | 100% (407) | Probable aminotransferase |
| M. tuberculosis H37Rv | Rv0075 | - | 3e-59 | 38.29% (410) | aminotransferase |
| M. tuberculosis H37Rv | Rv0858c | - | 3e-21 | 28.61% (381) | aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2316 | - | 0.0 | 100.00% (407) | aminotransferase |
| M. gilvum PYR-GCK | Mflv_1680 | - | 6e-22 | 27.33% (311) | aminotransferase |
| M. leprae Br4923 | MLBr_01794 | - | 0.0 | 76.17% (407) | aminotransferase |
| M. abscessus ATCC 19977 | MAB_0847c | - | 5e-26 | 30.50% (377) | aminotransferase |
| M. marinum M | MMAR_3453 | - | 0.0 | 78.52% (405) | aminotransferase |
| M. avium 104 | MAV_2132 | - | 0.0 | 78.66% (403) | transferase |
| M. smegmatis MC2 155 | MSMEG_5725 | - | 2e-20 | 28.01% (307) | aminotransferase |
| M. thermoresistible (build 8) | TH_1406 | - | 1e-147 | 67.25% (400) | Probable aminotransferase |
| M. ulcerans Agy99 | MUL_2727 | - | 0.0 | 77.78% (405) | aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_5070 | - | 9e-23 | 28.75% (313) | aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2294|M.tuberculosis_H37Rv MIP-----NPLEELTLEQLRSQRTSMKWRAHPADVLPLWVAEMDVKLPPT
Mb2316|M.bovis_AF2122/97 MIP-----NPLEELTLEQLRSQRTSMKWRAHPADVLPLWVAEMDVKLPPT
MMAR_3453|M.marinum_M MNP-----NPLEQLTLEQLRT-RTSMKWRAHPADVLPLWVAEMDVNLAPT
MUL_2727|M.ulcerans_Agy99 MNP-----NPLEQLTLEQLRT-RTSMKWRAHPADVLPLWVAEMDVNLAPT
MLBr_01794|M.leprae_Br4923 MTS-----NPLEQLTIEQLQC-RTSMKWRAHPEDVLPLWVAEMDVLLAPT
MAV_2132|M.avium_104 MTF-----NPFEELTLDQLRL-RTSMKWRAHPADVLPLWVAEMDVKLAPT
TH_1406|M.thermoresistible__bu MAPSAPVANPLEQLTLEQLRL-RTSMKWRVHPPDVLPLWVAEMDVPLADP
Mflv_1680|M.gilvum_PYR-GCK --------MTVQRLRPYAVTI-FAETSALAARVGAVNLGQGFPDEDGPPA
Mvan_5070|M.vanbaalenii_PYR-1 --------MTVRRLQPYAVTI-FAEMSALAARVGAVNLGQGFPDEDGPPA
MSMEG_5725|M.smegmatis_MC2_155 --------MTVRRLQPFAVTI-FAEMSALATRLGAVNLGQGFPDEDGPVE
MAB_0847c|M.abscessus_ATCC_199 -----MNQRTVERLRPFGATI-FAEMSALAVRHDAINLGQGFPDEDGPAS
....* :. . . ..: * . * .
Rv2294|M.tuberculosis_H37Rv VADALRRAIDDGDTGYP--YGT-EYAEAVREFACQRWQWHDLEVSRTAIV
Mb2316|M.bovis_AF2122/97 VADALRRAIDDGDTGYP--YGT-EYAEAVREFACQRWQWHDLEVSRTAIV
MMAR_3453|M.marinum_M VAAALHRAVDNGDTGYP--YGT-AYAEALAEFAALRWQWHDLEVGRTAIV
MUL_2727|M.ulcerans_Agy99 VAAALHRAVDNGDTGYP--YGT-AYAEALAEFAALRWQWHDLEVGRTAIV
MLBr_01794|M.leprae_Br4923 VAEAVHRAVDNGDTGYP--YGT-TFAEAISEFASQRWQWHDLDVSRTAIV
MAV_2132|M.avium_104 VAQALRTAIDHGDTGYP--CGT-AFAEAVSEFAARRWQWHDLDVSRTALV
TH_1406|M.thermoresistible__bu IAEALTTAVARGDTGYA--AGARRYAEAFAGFAARRWGWSGPAAEHTAIV
Mflv_1680|M.gilvum_PYR-GCK MLKAAENAIAEGVNQYPPGLGFAPLRQAIADQRRRRYGTVYDPDTEVLVT
Mvan_5070|M.vanbaalenii_PYR-1 MLKAAENAIAEGVNQYPPGLGIAPLRQAIADQRRRRYGSEYDPDTEVLVT
MSMEG_5725|M.smegmatis_MC2_155 MLKVAQNAIADGINQYPPGLGVPELRQAIAAQRKRVHGVQYDPDTEVLVT
MAB_0847c|M.abscessus_ATCC_199 MLDAAQQAIRSGLNQYPPGLGIPELRRAITADRLARYGEELDPDTQVLVT
: . *: * . *. * .*. .. :.
Rv2294|M.tuberculosis_H37Rv PDVMLGIVEVLRLITDRGDPVIVNSPVYAPFYAFVSHDGRRVIPAPLRGD
Mb2316|M.bovis_AF2122/97 PDVMLGIVEVLRLITDRGDPVIVNSPVYAPFYAFVSHDGRRVIPAPLRGD
MMAR_3453|M.marinum_M PDVMMGIVEMLRLVTDRGDTVIVNSPVYAPFYAFVSHDGRRVVEAPLGTD
MUL_2727|M.ulcerans_Agy99 PDVMMGIVEMLRLVTDRGDTVIVNSPVYAPFYAFVSHDGRRVVEAPLGTD
MLBr_01794|M.leprae_Br4923 PDVMLGVVEMLRLITDRGDGVIVSPPVYPPFYAFVTHDGRRVLEAPLGRD
MAV_2132|M.avium_104 PDVMLGAVEVLRLVTDPGDAVVVNPPVYAPFYAFVSHDGRRVVEAPLDDG
TH_1406|M.thermoresistible__bu PDVMLGIVEVLKLITDPGDAVVVNAPVYPPFYAFVTHAGRCVVEARLGAD
Mflv_1680|M.gilvum_PYR-GCK VGATEAIASSVLGLVEPGSEVLLIEPFYDSYSPVIAMAGCHRRAVPLRQN
Mvan_5070|M.vanbaalenii_PYR-1 VGATEAIAAAILGLVEPDSEVLLIEPFYDSYSPVIAMAGCHRRAVPMVRD
MSMEG_5725|M.smegmatis_MC2_155 VGATEAIAASVIGLVEPGSEVLVIEPFYDSYSPVIAMAGCERRAVPLVPS
MAB_0847c|M.abscessus_ATCC_199 VGATEAIAGALLGLVEPGSEVILIEPYYDSYAAVVAMAGAVRVPVPLVPD
.. . . : :.: .. *:: * * .: ..:: * . : .
Rv2294|M.tuberculosis_H37Rv GR---IDLDALQEAFSSARASSGSSGNVAYLLCNPHNPTGSVHTADELRG
Mb2316|M.bovis_AF2122/97 GR---IDLDALQEAFSSARASSGSSGNVAYLLCNPHNPTGSVHTADELRG
MMAR_3453|M.marinum_M GR---IDLDALEEAFAHARRAG---GNVAYLLCNPHNPTGAVHSYDELRG
MUL_2727|M.ulcerans_Agy99 GR---INLDALEEAFAHARRAG---GNVPYLLCNPHNPTGAAHSCDELRG
MLBr_01794|M.leprae_Br4923 GR---LDLAALQDAFSRARRSSGPNGKVAYLLCNPHNPTGSVHTVAELHG
MAV_2132|M.avium_104 GR---IDLGALEEAFAHARAGN--TAKVAYLLCNPHNPTGSVHTAAELRG
TH_1406|M.thermoresistible__bu GR---IDPASLEAAFGTAAGIG---DRAAYLLCSPHNPTGAVHTADELAH
Mflv_1680|M.gilvum_PYR-GCK GRGFAIDIEALRAAVGPR--------TRALIVNTPHNPTGMIASDEELRG
Mvan_5070|M.vanbaalenii_PYR-1 GRGFAIDVEGLRRAVTPK--------TRALILNSPHNPTGSVASDDELRA
MSMEG_5725|M.smegmatis_MC2_155 GRGFAIDVDALRAAVTPQ--------TKALILNSPHNPTGAVASDAELRA
MAB_0847c|M.abscessus_ATCC_199 GAGFALDTDALAAAITPR--------TAALVINSPHNPTGKVFTDTELAR
* :: .* *. . :: .****** : **
Rv2294|M.tuberculosis_H37Rv IAERAQRFGVRVVSDEIHAPLIPSGARFTPYLSVPGAEN-AFALMSASKA
Mb2316|M.bovis_AF2122/97 IAERAQRFGVRVVSDEIHAPLIPSGARFTPYLSVPGAEN-AFALMSASKA
MMAR_3453|M.marinum_M VAERARRYDVRVVSDEIHAPLVFSGARFTPYLTVPGSEN-ALSLTSATKA
MUL_2727|M.ulcerans_Agy99 VAERARRYDVRVVSDEIHAPLVFSGARFTPYLTVPGSEN-ALSLTSATKA
MLBr_01794|M.leprae_Br4923 VAELARRFGVRVISDEIHAPLVLSGARFTPYLSIPGAEN-AFALMSATKG
MAV_2132|M.avium_104 VAELARRFGVRVVSDEIHAPVILPGSRFTPFLTVAGAEN-AFALTSASKA
TH_1406|M.thermoresistible__bu VAALASRYGVRVISDEIHAPLVLPGASFTPYLSVPGAED-AFAVTSASKA
Mflv_1680|M.gilvum_PYR-GCK LAELAVAHDLLVITDEVYEHLVFDGRQHRPLAGYPGMAERTVTISSAGKM
Mvan_5070|M.vanbaalenii_PYR-1 VAELAISADLLVITDEVYEHLVFDGRRHRPLADYPGMAGRTITISSAGKM
MSMEG_5725|M.smegmatis_MC2_155 VAALAVEHDLLVITDEVYEALVFDGHRHLTLASYPGMRDRTVTISSAAKM
MAB_0847c|M.abscessus_ATCC_199 IAELAVEHDLLVVSDEVYERLLFDGRTQTPMASLPGMADRTVTISSAAKT
:* * .: *::**:: :: * . .* :.:: ** *
Rv2294|M.tuberculosis_H37Rv WNLGGLKAALAIAGREAAADLARMPEEVGHGPSHLGVIAHTAAFRTGGNW
Mb2316|M.bovis_AF2122/97 WNLGGLKAALAIAGREAAADLARMPEEVGHGPSHLGVIAHTAAFRTGGNW
MMAR_3453|M.marinum_M WNLGGVKAALAVAGPEAAADLRRMPEEVSHGPSHLGIIAHTEAFRTGGDW
MUL_2727|M.ulcerans_Agy99 WNLGGVKAALAVAGPEAAADLRRMPEEVSHGPSHLGIIAHTEAFRTGGDW
MLBr_01794|M.leprae_Br4923 WNLCGLKAAIAIAGREAAADLRRIPKEVSHGPSHLGIIAHTAAFRTGSSW
MAV_2132|M.avium_104 WNLSGLKAALAIAGPEAAADLNRMPEEVSHGPSHLGVIAHTEAFRSGDGW
TH_1406|M.thermoresistible__bu WNLAGLKAGLVIAGPDAAADLDRMPEEVSHGPSHLGVIAHTVSFAEGEPW
Mflv_1680|M.gilvum_PYR-GCK FNVTGWKIGWACGPSDLIAGVRAAKQYLSYVAGAPFQPAVAQALNNEDAW
Mvan_5070|M.vanbaalenii_PYR-1 FNVTGWKIGWACGPSDLIAGVRAAKQYLSYVGGAPFQPAVAHALATEDAW
MSMEG_5725|M.smegmatis_MC2_155 FNATGWKIGWACGAPELIAGVRAAKQYLTYVGGAPFQPAVAHALNHEDGW
MAB_0847c|M.abscessus_ATCC_199 FNCTGWKIGWACGTPELVAGVRAAKQFLSYVGGGPFQPAVAVALDTEQSW
:* * * . . . : *.: : : : . * : :: *
Rv2294|M.tuberculosis_H37Rv LDALLRGLDHNRTLLGALVDEHLPGVQYRWPQGTYLAWLDCRELGFDDAA
Mb2316|M.bovis_AF2122/97 LDALLRGLDHNRTLLGALVDEHLPGVQYRWPQGTYLAWLDCRELGFDDAA
MMAR_3453|M.marinum_M LDALLAGLDANRTLLSDLVDAQLPGVRLRWPEGTYLAWLDCRDLGISEQD
MUL_2727|M.ulcerans_Agy99 LDALLAGLDANRTLSSDLVDAQLPGVRLRWPQGTYLAWLDCRDLGISEQD
MLBr_01794|M.leprae_Br4923 LDALLQGLEANRALLRDLVTKHLPGVRYQRPQGTYLTWLDCRHLGFDEQI
MAV_2132|M.avium_104 LDATLQGLDQNRALLGELVAEHLPGVKYQWPQGTYLAWLDCRELGFAEDA
TH_1406|M.thermoresistible__bu LDALLAGLAANRDLLGRLLAERLPLVGYTPPQGTYLAWLDCRRLGLDEAG
Mflv_1680|M.gilvum_PYR-GCK VDALCASFQARRDRLSSALSD--IGFDVFDSFGTYFLTVDPRPLGYDDST
Mvan_5070|M.vanbaalenii_PYR-1 VDALCASFQSRRDRLGSALSD--IGFEVHESFGTYFLCVDPRPLGFTDSA
MSMEG_5725|M.smegmatis_MC2_155 VADLRDSFQSKRDRLGSALAE--IGFEVHDSRGTYFLCADPRPLGYDDST
MAB_0847c|M.abscessus_ATCC_199 VKTLRESLQRRRDRLSAALTG--LGFEVHSSDGGYFVCADPRPLGFSDSA
: .: .* : . . * *: * * ** :
Rv2294|M.tuberculosis_H37Rv SDEMTEGLAVVSDLSGPARWFLDHARVALSSGHVFGIGGAGHVRINFATS
Mb2316|M.bovis_AF2122/97 SDEMTEGLAVVSDLSGPARWFLDHARVALSSGHVFGIGGAGHVRINFATS
MMAR_3453|M.marinum_M ----ASGLAVVSDLSGPASWFLDHARVALSSGHVFGTGGDGHVRMNFATS
MUL_2727|M.ulcerans_Agy99 ----ASGLAVVSDLSGPASWFLDHARVALSSGHVFGTGGDGHVRMNFATS
MLBr_01794|M.leprae_Br4923 N----EGLAAISDLSGPARWFLDHSRVALTSGHAFGTGGVGHVRVNFATS
MAV_2132|M.avium_104 ----AQGLAVVADLSGPARWFLEHARVALSSGHVFGTGGAGHLRLNFATS
TH_1406|M.thermoresistible__bu G---GDGPGIVSELAGPARVFLDQARVALSSGHVFGSGGAGHVRLNFATS
Mflv_1680|M.gilvum_PYR-GCK --------SFCAQLPEQVGVAAIPMSAFCDPDAAHAGEWKHLVRFAFCKR
Mvan_5070|M.vanbaalenii_PYR-1 --------AFCAELPERVGVAAIPMSAFCDPAAGHAEEWKHLVRFAFCKR
MSMEG_5725|M.smegmatis_MC2_155 --------AFCAELPTRAGVAAIPMSAFCDPDAPHADVWNHLVRFAFCKR
MAB_0847c|M.abscessus_ATCC_199 --------ELCRRLPETVGVAAVPVSAFCD---RYADQWNHLVRFTFAKR
*. . . .. :*. *..
Rv2294|M.tuberculosis_H37Rv RAILIEAVSRMSRSLLERR-
Mb2316|M.bovis_AF2122/97 RAILIEAVSRMSRSLLERR-
MMAR_3453|M.marinum_M QAILTEAVTRMGKALGELR-
MUL_2727|M.ulcerans_Agy99 QAILTEAVTRMGKALGELR-
MLBr_01794|M.leprae_Br4923 QAILIEALSRMGRALVNFR-
MAV_2132|M.avium_104 RAILTEALSRMGRALAARSG
TH_1406|M.thermoresistible__bu QAILTEAITRMSTVRAARCE
Mflv_1680|M.gilvum_PYR-GCK DETLDEAIRRLQALRSTG--
Mvan_5070|M.vanbaalenii_PYR-1 EDTLDEAIRRLRALPRSR--
MSMEG_5725|M.smegmatis_MC2_155 DDTLDEAIRRLQALRRC---
MAB_0847c|M.abscessus_ATCC_199 DVVIDEAITRLARLGA----
: **: *: