For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VREISVPAPFTVGEHDNVAAMVFEHERDDPDYVIYQRLIDGVWTDVTCAEAANQIRAAALGLISLGVQAG DRVVIFSATRYEWAILDFAILAVGAVTVPTYETSSAEQVRWVLQDSEAVVLFAETDSHATMVAELSGSVP ALREVLQIAGSGPNALDRLTEAGASVDPAELTARLAALRSTDPATLIYTSGTTGRPKGCQLTQSNLVHEI KGARAYHPTLLRKGERLLVFLPLAHVLARAISMAAFHSKVTVGFTSDIKNLLPMLAVFKPTVVVSVPRVF EKVYNTAEQNAANAGKGRIFAIAAQTAVDWSEACDRGGPGLLLRAKHAVFDRLVYRKLRAALGGNCRAAV SGGAPLGARLGHFYRGAGLTIYEGYGLSGTSGGVAISQFNDLKIGTVGKPVPGNSLRIADDGELLVRGGV VFSGYWRNEQATTEAFTDGWFKTGDLGAVDEDGFLTITGRKKEIIVTAGGKNVAPAVLEDQLRAHPLISQ AVVVGDAKPFIGALITIDPEAFEGWKQRNSKTAGASVGDLATDPDLIAEIDAAVKQANLAVSHAESIRKF RILPVDFTEDTGELTPTMKVKRKVVAEKFASDIEAIYNKE
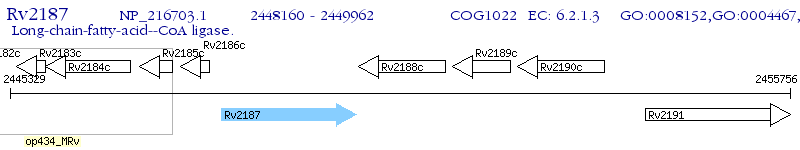
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2187 | fadD15 | - | 100% (600) | Probable long-chain-fatty-acid-CoA ligase fadD15 (FATTY-ACID-CoA SYNTHETASE) (FATTY-ACID-CoA SYNTHASE) |
| M. tuberculosis H37Rv | Rv1550 | fadD11 | 5e-66 | 37.42% (449) | fatty-acid-CoA ligase |
| M. tuberculosis H37Rv | Rv0166 | fadD5 | 4e-32 | 28.05% (467) | acyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2210 | fadD15 | 0.0 | 99.61% (508) | long-chain-fatty-acid-CoA ligase fadD15 (fatty-acid-CoA |
| M. gilvum PYR-GCK | Mflv_2962 | - | 0.0 | 71.24% (598) | AMP-dependent synthetase and ligase |
| M. leprae Br4923 | MLBr_00887 | - | 0.0 | 80.50% (600) | putative long-chain-fatty-acid-CoA ligase |
| M. abscessus ATCC 19977 | MAB_1978c | - | 0.0 | 70.57% (598) | long-chain-fatty-acid--CoA ligase FadD |
| M. marinum M | MMAR_3231 | fadD15 | 0.0 | 82.17% (600) | long-chain-fatty-acid-CoA ligase FadD15 |
| M. avium 104 | MAV_2307 | - | 0.0 | 80.50% (600) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4254 | - | 0.0 | 73.08% (598) | AMP-binding enzyme |
| M. thermoresistible (build 8) | TH_3481 | - | 0.0 | 71.24% (598) | PUTATIVE AMP-dependent synthetase and ligase |
| M. ulcerans Agy99 | MUL_3542 | fadD15 | 0.0 | 81.83% (600) | long-chain-fatty-acid-CoA ligase FadD15 |
| M. vanbaalenii PYR-1 | Mvan_3549 | - | 0.0 | 71.33% (600) | AMP-dependent synthetase and ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2962|M.gilvum_PYR-GCK MTAREFTVPAPFTVEERDNIVGAVFTHERDDPDHVIFQRLVDGKWIDVTA
MSMEG_4254|M.smegmatis_MC2_155 --MREDSIPASFTVGEYDNVAGCVYAHETDDPDHVILRRLVDGTWTDVTC
TH_3481|M.thermoresistible__bu --VREFHEPATFTVGERDNIVSAVFDHERDDPQHVIFRRRIDGRWTDVTC
Mvan_3549|M.vanbaalenii_PYR-1 --MREFSVPATFTVGENDNVVSTVYSHERDDPDHVIFQRLVNGTWVDVTC
Rv2187|M.tuberculosis_H37Rv --MREISVPAPFTVGEHDNVAAMVFEHERDDPDYVIYQRLIDGVWTDVTC
Mb2210|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00887|M.leprae_Br4923 --MREFSVPTRFSVGEHDNIAAVVFEHERDDPNLVIYQRQIDGVWTDVTC
MAV_2307|M.avium_104 --MREYSVPARFSVGENENIATMVFEHERDDPNFVIYQRQVDGEWTDVTC
MMAR_3231|M.marinum_M --MRQYSVPARFSVDERDNVAAMVFEHERDDPDHIIYQRQIDGIWTDITC
MUL_3542|M.ulcerans_Agy99 --MRQYSVPARFSVDERDNVAAMVFEHERDDPDHIIYQRQIDGIWTDITC
MAB_1978c|M.abscessus_ATCC_199 --MREFSVPAPFTVEDNASVVRAVYDYEREDPNQAAFSRLIDDTWTPVTY
Mflv_2962|M.gilvum_PYR-GCK RQAAGQIRSAARGLIAEGVQAGDRVALLSATRYEWPILDFAILSIGAVTV
MSMEG_4254|M.smegmatis_MC2_155 AQAAAQIRSAALGLIAEGVQPGDRVAILSATRYEWPIIDFAILSIGAVTV
TH_3481|M.thermoresistible__bu AQAAEQVRAAALGLIAEGVQPGDRVALMSATRYEWVILDYAILAVGALTV
Mvan_3549|M.vanbaalenii_PYR-1 RQAAEQIRSVASGLIAEGVQPGDRVALLSATRYEWPILDYAILSVGAVTV
Rv2187|M.tuberculosis_H37Rv AEAANQIRAAALGLISLGVQAGDRVVIFSATRYEWAILDFAILAVGAVTV
Mb2210|M.bovis_AF2122/97 --------------------------------------------MGAVTV
MLBr_00887|M.leprae_Br4923 AEAASQIRSAALGLIALGMQAGDRVSIFSATCFEWAILDLAILSVGAVTV
MAV_2307|M.avium_104 AEAAAQIRSAALGLISLGVQAGDRVSIFSATRYEWAILDLAILAVGAVTV
MMAR_3231|M.marinum_M AEAARHIRSAALGLIALGVQAGDRVSIFSATCYEWAILDLAILAVGAVTV
MUL_3542|M.ulcerans_Agy99 AEAARHIRSAALGLIALGVQASDRVSIFSATCYEWAILDLAILAVGAVTV
MAB_1978c|M.abscessus_ATCC_199 AEAAAQIRAVANGLIAKGVAPGDRVALISATRYEWTIIDYAVLSIGAITV
:**:**
Mflv_2962|M.gilvum_PYR-GCK PIYETSSAEQVQFVLADSGAVAAIAETDAHAAHIEAQRDTLPALRRIYVI
MSMEG_4254|M.smegmatis_MC2_155 PIYETSAAEQVRFVLENSESVLVFAETDAHADKVEQLRDQLPALRKVFRI
TH_3481|M.thermoresistible__bu PIFETSSADQVRWVLQDSGAVLAVVETEKHGQLVKSVQDDAPALRKTLLI
Mvan_3549|M.vanbaalenii_PYR-1 PIYETSSPEQVRWVLEDSGAVLAFVELEAHALMVKELLNELPALRKVMVI
Rv2187|M.tuberculosis_H37Rv PTYETSSAEQVRWVLQDSEAVVLFAETDSHATMVAELSGSVPALREVLQI
Mb2210|M.bovis_AF2122/97 PIYETSSAEQVRWVLQDSEAVVLFAETDSHATMVAELSGSVPALREVLQI
MLBr_00887|M.leprae_Br4923 PIYETSSAKQVRWILQDSDAVLVFAETDAHATMVNQLAGELPALRQVLQI
MAV_2307|M.avium_104 PIYETSSAEQVRWVLQDSEAVLAFAETDEHAAMVTELTAELPALRRVLHI
MMAR_3231|M.marinum_M PIYETSSAEQVRWVLQNSEAVLAFAETDAHAAMIAELTGDLPALRRVLVI
MUL_3542|M.ulcerans_Agy99 PIYETSSAEQVRWVLQNSEAVLAFAETDAHAAMIAELTGDLPALRRVLVI
MAB_1978c|M.abscessus_ATCC_199 PIYETSSADQVRWVLQDSAAVALFVESDAHAKIAEELAPELPNLRTVFHI
* :***:..**:::* :* :* ..* : *. * ** *
Mflv_2962|M.gilvum_PYR-GCK D-GS-VPALDELAEAGASVDAAAVDERVASIRSTDAATLIYTSGTTGRPK
MSMEG_4254|M.smegmatis_MC2_155 D-GSGTPALDELAEAGKDVDPAELDKRLANIKSSDPATLIYTSGTTGQPK
TH_3481|M.thermoresistible__bu E-GSGPTAIDELTAAGSGVDRAELDNRLAGIRAADPATLIYTSGTTGRPK
Mvan_3549|M.vanbaalenii_PYR-1 E-TAAPSALDALAEAGATVDPGEVDRRVAAIRSADPATLIYTSGTTGRPK
Rv2187|M.tuberculosis_H37Rv A-GSGPNALDRLTEAGASVDPAELTARLAALRSTDPATLIYTSGTTGRPK
Mb2210|M.bovis_AF2122/97 A-GSGPNALDRLTEAGASVDPAELTARLAALRSTDPATLIYTSGTTGRPK
MLBr_00887|M.leprae_Br4923 N-SAGATALDQLAEAGASVDVAQLANRLKTLRAQDPATLIYTSGTTGRPK
MAV_2307|M.avium_104 D-GSGPKALDQLAEAAAGVEPAELTARLDGLRAQDPATLIYTSGTTGRPK
MMAR_3231|M.marinum_M N-GSGPKALEQLAEEGGSVDRAELTARLDALRSSDPATLIYTSGTTGRPK
MUL_3542|M.ulcerans_Agy99 N-GSGPKALEQLAEEGGSVDRAELTARLDALRSSDRATLIYTSGTTGRPK
MAB_1978c|M.abscessus_ATCC_199 ASTSAPAALDELAEAGAGVDPAELEGRLAAIKATDPATLIYTSGTTGRPK
: *:: *: . *: . : *: ::: * ***********:**
Mflv_2962|M.gilvum_PYR-GCK GCQLTHSNLLYEIRGAKACFPEHLAKGEKLLVFLPLAHVLARALTIGAFS
MSMEG_4254|M.smegmatis_MC2_155 GCQLTHSNLLHEIRGQKECFPDHLAKGERILVFLPLAHVLARAITIGAFA
TH_3481|M.thermoresistible__bu GCVLTHSNLVHEVRADRDCFPTLLDKGQRMLTFLPLAHVLARAISMAAFH
Mvan_3549|M.vanbaalenii_PYR-1 GCQLTHSNLLYETRGATACFPGLLRKGERLLVFLPLAHVLARALSMTAFA
Rv2187|M.tuberculosis_H37Rv GCQLTQSNLVHEIKGARAYHPTLLRKGERLLVFLPLAHVLARAISMAAFH
Mb2210|M.bovis_AF2122/97 GCQLTQSNLVHEIKGARAYHPTLLRKGERLLVFLPLAHVLARAISMAAFH
MLBr_00887|M.leprae_Br4923 GCQLTHSNLLYEVRGAKECLPTLLQPGQRLLVFLPLAHVLARALTLSAFT
MAV_2307|M.avium_104 GCQLTHSNLLYETRGAKESLPTLLDQGQRLLIFLPLAHVLARSLTLSAFA
MMAR_3231|M.marinum_M GCQLTHSNLLHEIRGTQECLPTLLTPGQRLLVFLPLAHVLARALTLSAFA
MUL_3542|M.ulcerans_Agy99 GCQLTHSNLLHEIRGTQECLPTLLTPGQRLLVFLPLAHVLARALTLSAFA
MAB_1978c|M.abscessus_ATCC_199 GCQLTHSNLVYETRGARAVFPDYLQKGSRTLIFLPLAHVLARGIAMACFQ
** **:***::* :. * * *.: * **********.::: .*
Mflv_2962|M.gilvum_PYR-GCK NGVTLGFTSDIKNLVPMFAVFQPTLVVSVPRVFEKVYNTAELNAETGGKG
MSMEG_4254|M.smegmatis_MC2_155 NKVTLGFTSDIKNLVPMFGVFKPTLVISVPRVFEKVYNTAEQNARNDGKG
TH_3481|M.thermoresistible__bu SKVTQGFTSDLANLVPLFGEFKPTAVVSVPRVFEKVYNTAELNAQEGGKG
Mvan_3549|M.vanbaalenii_PYR-1 NGVTIGYTSDIKNLVPMFGQFKPTVIVSVPRVFEKVYNTAELNAQDSGKG
Rv2187|M.tuberculosis_H37Rv SKVTVGFTSDIKNLLPMLAVFKPTVVVSVPRVFEKVYNTAEQNAANAGKG
Mb2210|M.bovis_AF2122/97 SKVTVGFTSDIKNLLPMLAVFKPTVVVSVPRVFEKVYNTAEQNAANAGKG
MLBr_00887|M.leprae_Br4923 YKMTVGFTSDIKNLLPMLAVFKPTVVVSVPRVFEKVYNTAEQNAANEGKS
MAV_2307|M.avium_104 NKVTVGFTSDIKNLLPMFAVFKPTVVVSVPRVFEKVYNTAEQNAANDGKG
MMAR_3231|M.marinum_M SKVTVGFTSDIKNLLPLFAVFKPTVVVSVPRVFEKVYNTAEQNASNDGKG
MUL_3542|M.ulcerans_Agy99 SKVTVGFTSDIKNLLPLFAVFKPTVVVSVPRVFEKVYNTAEQNASNDGKG
MAB_1978c|M.abscessus_ATCC_199 SKVSVGFTSDIKTLVPTFGIFKPTFIVSVPRIFEKVYNTARQNAQNDGKG
:: *:***: .*:* :. *:** ::****:********. ** **.
Mflv_2962|M.gilvum_PYR-GCK KIFTAAANTAIEYSKALDSGGPGLVLKLKHALFDKLVYGKLRAALGGRCH
MSMEG_4254|M.smegmatis_MC2_155 KIFEIAAETAIEFSKAQDKGGPGLLLRVKHAVFDKLVYGKLRAALGGECH
TH_3481|M.thermoresistible__bu AIFAQAARTAIDWSKALDTGGPGLWLRAKHALFDKLVYGKLRAALGGECR
Mvan_3549|M.vanbaalenii_PYR-1 AIFAMAVQTAIDWSMAQDTGGPGLLLRAKHALFDRLVYGKLRAATGGDCR
Rv2187|M.tuberculosis_H37Rv RIFAIAAQTAVDWSEACDRGGPGLLLRAKHAVFDRLVYRKLRAALGGNCR
Mb2210|M.bovis_AF2122/97 RIFAIAAQTAVDWSEACDRGGPGLLLRAKHAVFDRLVYRKLRAALGGNCR
MLBr_00887|M.leprae_Br4923 RIFAMAAQTAVEWSEASHRGGPGLLLRARHAVFDRLVYRKLRAALGGDCH
MAV_2307|M.avium_104 RIFAMAVQTAVDWSEAADRGRRGPLLRAKHALFDRLVYHKLRAALGGDCH
MMAR_3231|M.marinum_M AIFKLAAQTAVDWSRAWDDGRPGLLLRAKHALFDRLVYHKLRAALGGDCH
MUL_3542|M.ulcerans_Agy99 AIFKLAAQTAVDWSRAWDDGRPGLLLRAKHALFDRLVYHKLRAALGGDCH
MAB_1978c|M.abscessus_ATCC_199 KIFDAAADTAIEYSEALDKGGPGIVLRAKRAAFDKLVYGKLRAALGGECV
** *. **:::* * . * * *: ::* **:*** ***** ** *
Mflv_2962|M.gilvum_PYR-GCK AAISGGAPLGARLGHFYRGVGLSIYEGYGLTETSAAITVNRVGDLRVGSV
MSMEG_4254|M.smegmatis_MC2_155 AAISGGAPLGERLGHFYRGVGLSIYEGYGLTETSAAITVNRLNDLKVGSV
TH_3481|M.thermoresistible__bu AAISGGAPLGARLGHFFRGAGVTVYEGYGLTETSAAITVNRMDDLRIGTV
Mvan_3549|M.vanbaalenii_PYR-1 ASISGGAPLGARLGHFFRGVGLTIYEGYGMTETSAAITANRIGELKVGSV
Rv2187|M.tuberculosis_H37Rv AAVSGGAPLGARLGHFYRGAGLTIYEGYGLSGTSGGVAISQFNDLKIGTV
Mb2210|M.bovis_AF2122/97 AAVSGGAPLGARLGHFYRGAGLTIYEGYGLSETSGGVAISQFNDLKIGTV
MLBr_00887|M.leprae_Br4923 ASVSGGAPLGARLGHFYRGVGLTIHEGYGLTETSAAITLNQAGNVKVGTV
MAV_2307|M.avium_104 ASVSGGAPLGSRLGHFFRGVGLTIHEGYGLTETSSAITVNQVGNVKIGTV
MMAR_3231|M.marinum_M AAVSGGAPLGARLGHFYRGVGLTIYEGYGLTETSAAVTVNQIDALKIGTV
MUL_3542|M.ulcerans_Agy99 AAVSGGAPLGARLGHFYRGVGLTIYEGYGLTETSAAVTVNQIDALKIGTV
MAB_1978c|M.abscessus_ATCC_199 ASISGGAPLGARLGHFYRGVGLTIYEGYGLTETTAACAVNVIGGLKIGSV
*::******* *****:**.*::::****:: *:.. : . . :::*:*
Mflv_2962|M.gilvum_PYR-GCK GKLVPGNSMRLGEDGELLVRGGVVFNGYWGNEEETRAVFT-DGWFHTGDL
MSMEG_4254|M.smegmatis_MC2_155 GRLVPGNSMRIADDGELLVKGGVVFNGYWKNEDETKAVIDADGWFHTGDL
TH_3481|M.thermoresistible__bu GKLVPGCSMKLAPDGELLVKGGVVFNGYWQNEEATKEAIV-DGWFHTGDL
Mvan_3549|M.vanbaalenii_PYR-1 GKLLPGNSMAIANDGELLVRGGVVFNGYWRNEKATRETVV-NGWLHTGDL
Rv2187|M.tuberculosis_H37Rv GKPVPGNSLRIADDGELLVRGGVVFSGYWRNEQATTEAFT-DGWFKTGDL
Mb2210|M.bovis_AF2122/97 GKPVPGNSLRIADDGELLVRGGVVFSGYWRNEQATTEAFT-DGWFKTGDL
MLBr_00887|M.leprae_Br4923 GNLLPGNSMRIADDGELLVRGNVVFSGYWHNEQGTIEAFT-EGWFKTGDL
MAV_2307|M.avium_104 GTLLPGNSMRIAEDGELLVRGGVVFSGYWRNEQATADAFT-DGWFKTGDL
MMAR_3231|M.marinum_M GKLVPGNSLRIADDGELLVRGGVVFSGYWRNEQATDEAFT-DGWFRTGDL
MUL_3542|M.ulcerans_Agy99 GKLVPGNSLRIADDGELLVRGGVVFSGYWRNEQATDEAFT-DGWFRTGDL
MAB_1978c|M.abscessus_ATCC_199 GRPLPGNAVKIAEDGELLVSGGVVFSGYWNNETATSESIV-DGWFHTGDL
* :** :: :. ****** *.***.*** ** * . :**::****
Mflv_2962|M.gilvum_PYR-GCK GAIDDDGFLTIVGRKKEIIVTAGGKNVAPAQLEDRLRAHPLISQAMAVGD
MSMEG_4254|M.smegmatis_MC2_155 GAIDDNGFLTIVGRKKEIIVTAGGKNVAPALLEDRLRAHPLISQAMAVGD
TH_3481|M.thermoresistible__bu GSIDDDGFLTIVGRKKEIIVTAGGKNVAPAVLEDQLRAHPLISQAMAVGD
Mvan_3549|M.vanbaalenii_PYR-1 GSIDEDGYLSIVGRKKEIIVTAGGKNVAPAVLEDQLRAHPLISQAMAVGD
Rv2187|M.tuberculosis_H37Rv GAVDEDGFLTITGRKKEIIVTAGGKNVAPAVLEDQLRAHPLISQAVVVGD
Mb2210|M.bovis_AF2122/97 GAVDEDGFLTITGRKKEIIVTAGGKNVAPAVLEDQLRAHPLISQAVVVGD
MLBr_00887|M.leprae_Br4923 GAIDEDGFLTITGRKKEIIVTAGGKNVAPALLEDQLRAHPLISQALVVGD
MAV_2307|M.avium_104 GVLDDDGFLKIVGRKKELIVTAGGKNVAPAVLEDRLRAHPLISQAMVVGD
MMAR_3231|M.marinum_M GAIDDDGFLSITGRKKELIVTAGGKNVAPAVLEDQLRAHPLISQAMVVGD
MUL_3542|M.ulcerans_Agy99 GAIDDDGFLSITGRKKELIVTAGGKNVAPAVLEDQLRAHPLISQAVVVGD
MAB_1978c|M.abscessus_ATCC_199 GAIDEDGFVTITGRKKEIIVTAGGKNVAPAVLEDQLRAHPLISQAMAVGD
* :*::*::.*.*****:************ ***:**********:.***
Mflv_2962|M.gilvum_PYR-GCK QQPFIAALITVDTEAFPGWKERHHKASDASVADLADDPDLLAEIELAVKD
MSMEG_4254|M.smegmatis_MC2_155 KQPFIAALITIDPEAFPAWKERNAKSASATVAELAEDPDLIAEINLAVKE
TH_3481|M.thermoresistible__bu QKPFIGALITIDPEAFETWKQHRGKNPDATLAEMTEDPDLVAEIDEAIQQ
Mvan_3549|M.vanbaalenii_PYR-1 AKPFIGALIAIDTEAFEVWKQHHGKAPAASVAELIDDPDLVGEIDLAVKS
Rv2187|M.tuberculosis_H37Rv AKPFIGALITIDPEAFEGWKQRNSKTAGASVGDLATDPDLIAEIDAAVKQ
Mb2210|M.bovis_AF2122/97 AKPFIGALITIDPEAFEGWKQRNSKTAGASVGDLATDPDLIAEIDAAVKQ
MLBr_00887|M.leprae_Br4923 AKPFVGALITIDPEAFNGWKQRNSKAADAAIGDLTTDPDLVAEVDAAVKQ
MAV_2307|M.avium_104 NKPFIGALITIDPEAFGGWKQRNHKGAGASVADLTTDPDLVAEVDAAIKD
MMAR_3231|M.marinum_M AKPFIGALITIDPEAFGGWKQRNSKADHAAVRDLAEDPDLVAEVDAAVKE
MUL_3542|M.ulcerans_Agy99 AKPFIGALITIDPEAFGGWKQRNSKADHAAVRDLAEDPDLVAGVDAAVKE
MAB_1978c|M.abscessus_ATCC_199 KQPFIGALITIDPEAIDGWKQRNGKPADATVADLVEDSDLLAEIGTAVKA
:**:.***::*.**: **::. * *:: :: *.**:. : *::
Mflv_2962|M.gilvum_PYR-GCK ANQAVSKAEAIRKFRILPVDFTEDTGELTPTLKVKRKVVAEKFAADIEAL
MSMEG_4254|M.smegmatis_MC2_155 ANQAVSNAEAIRKFRILPVDFTEDTGELTPTLKVKRKVVAEKFATDIAAL
TH_3481|M.thermoresistible__bu ANKAVSRAEAIRKFRILPVDFTVEDGELTPTLKLKRAVVAEKYAAEIEKI
Mvan_3549|M.vanbaalenii_PYR-1 ANQAVSKAEAIRKFRILPVDFTVFTGELTPTLKVRRNVVADKFAEEIERL
Rv2187|M.tuberculosis_H37Rv ANLAVSHAESIRKFRILPVDFTEDTGELTPTMKVKRKVVAEKFASDIEAI
Mb2210|M.bovis_AF2122/97 ANLAVSHAESIRKFRILPVDFTEDTGELTPTMKVKRKVVAEKFASDIEAI
MLBr_00887|M.leprae_Br4923 ANLSVSHAESIRKFRILPVDFTEQTGELTPTMKVKRNVVAQKFASDIEAI
MAV_2307|M.avium_104 ANQAVSHAESIRKFRILPVDFTEDTGELTPTMKVKRNVVAEKFASDIEAI
MMAR_3231|M.marinum_M ANLAVSHAESIRKFRILHVDFTEDTGELTPTMKVKRNVVAEKFSVEIEAI
MUL_3542|M.ulcerans_Agy99 ANLAVSHAESIRKFRILHVDFTEDTGELTPTMKVKRNVVAEKFSVEIEAI
MAB_1978c|M.abscessus_ATCC_199 ANQVVSHAEAIKKFRILPVDFTEATGEMTPTLKVKRNVVAEKFASDIDAI
** **.**:*:***** **** **:***:*::* ***:*:: :* :
Mflv_2962|M.gilvum_PYR-GCK YAKG--
MSMEG_4254|M.smegmatis_MC2_155 YA----
TH_3481|M.thermoresistible__bu YSS---
Mvan_3549|M.vanbaalenii_PYR-1 YTTDSA
Rv2187|M.tuberculosis_H37Rv YNKE--
Mb2210|M.bovis_AF2122/97 YNKE--
MLBr_00887|M.leprae_Br4923 YEKD--
MAV_2307|M.avium_104 YAKD--
MMAR_3231|M.marinum_M YTKD--
MUL_3542|M.ulcerans_Agy99 YTKD--
MAB_1978c|M.abscessus_ATCC_199 YAGAPQ
*