For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSARGTLWGVGLGPGDPELVTVKAARVIGEADVVAYHSAPHGHSIARGIAEPYLRPGQLEEHLVYPVTTE ATNHPGGYAGALEDFYADATERIATHLDAGRNVALLAEGDPLFYSSYMHLHTRLTRRFNAVIVPGVTSVS AASAAVATPLVAGDQVLSVLPGTLPVGELTRRLADADAAVVVKLGRSYHNVREALSASGLLGDAFYVERA STAGQRVLPAADVDETSVPYFSLAMLPGGRRRALLTGTVAVVGLGPGDSDWMTPQSRRELAAATDLIGYR GYLDRVEVRDGQRRHPSDNTDEPARARLACSLADQGRAVAVVSSGDPGVFAMATAVLEEAEQWPGVRVRV IPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWDVIAARLTAAAAADLVLAIYNPASVTRTWQVGAMRE LLLAHRDPGIPVVIGRNVSGPVSGPNEDVRVVKLADLNPAEIDMRCLLIVGSSQTRWYSVDSQDRVFTPR RYPEAGRATATKSSRHSD
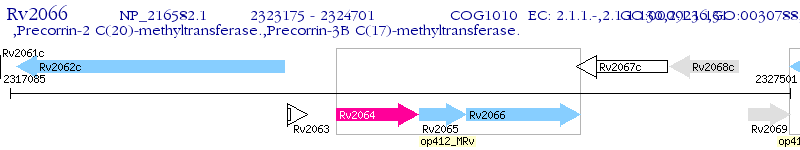
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2066 | cobI | - | 100% (508) | Probable bifunctional protein, CobI-CobJ fusion protein: S-adenosyl-L-methionine-precorrin-2 methyl transferase + precorrin-3 methylase |
| M. tuberculosis H37Rv | Rv2847c | cysG | 2e-08 | 31.33% (166) | multifunctional uroporphyrinogen III methylase/precorrin-2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2092 | cobI | 0.0 | 100.00% (508) | bifunctional S-adenosyl-L-methionine-precorrin-2 methyl |
| M. gilvum PYR-GCK | Mflv_3104 | - | 0.0 | 80.61% (495) | precorrin-3B C17-methyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2198c | - | 0.0 | 72.06% (494) | cobalamin biosynthesis protein CobI |
| M. marinum M | MMAR_3046 | cobI | 0.0 | 88.64% (493) | bifunctional protein: CobI-CobJ fusion protein |
| M. avium 104 | MAV_2430 | - | 0.0 | 86.15% (491) | cobalamin biosynthesis protein cobIJ |
| M. smegmatis MC2 155 | MSMEG_3873 | - | 0.0 | 79.23% (496) | cobalamin biosynthesis protein cobIJ |
| M. thermoresistible (build 8) | TH_0440 | cobI | 0.0 | 82.11% (492) | Probable bifunctional protein, CobI-CobJ fusion protein: |
| M. ulcerans Agy99 | MUL_2290 | cobI | 0.0 | 88.03% (493) | bifunctional protein, CobI-CobJ fusion protein |
| M. vanbaalenii PYR-1 | Mvan_3431 | - | 0.0 | 79.84% (491) | precorrin-3B C17-methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2066|M.tuberculosis_H37Rv MSA--------RGTLWGVGLGPGDPELVTVKAARVIGEADVVAYHSAPHG
Mb2092|M.bovis_AF2122/97 MSA--------RGTLWGVGLGPGDPELVTVKAARVIGEADVVAYHSAPHG
MMAR_3046|M.marinum_M MTD--------RGTLWGVGLGPGDPELVTVKAARVIGEADVVAYHSARHG
MUL_2290|M.ulcerans_Agy99 MTD--------RGTLWGVGLGPGDPELVTVKAARVIGEADVVAYHSARHG
MAV_2430|M.avium_104 MTAPANP----RGTLWGVGLGPGDPELVTVKAARVIGEADVVAYHSARHG
MSMEG_3873|M.smegmatis_MC2_155 MSENKHG------TLYGVGLGPGDPELVTVKAARVIGEADVVAYHSARHG
TH_0440|M.thermoresistible__bu MTVSQHGGAQHRGTLYGVGLGPGDPELVTVKAARVIGEADVVAYHSARHG
Mflv_3104|M.gilvum_PYR-GCK MSARQ-------GTLWGVGLGPGDPELVTVKAARVIGAADVIAYHSARHG
Mvan_3431|M.vanbaalenii_PYR-1 MTGSQEP----KGTLWGVGLGPGDPELVTVKAARVIGAADVVAYHSARHG
MAB_2198c|M.abscessus_ATCC_199 MTDANKP----RGVLWGVGLGPGDPELVTVKAARVIGDADVVAFHSARHG
*: .*:********************* ***:*:*** **
Rv2066|M.tuberculosis_H37Rv HSIARGIAEPYLRPGQLEEHLVYPVTTEATNHPGGYAGALEDFYADATER
Mb2092|M.bovis_AF2122/97 HSIARGIAEPYLRPGQLEEHLVYPVTTEATNHPGGYAGALEDFYADATER
MMAR_3046|M.marinum_M RSIARGIAERYLRPGQIEEHLIYPVTTETTDHPGGYAGALEDFYEQATER
MUL_2290|M.ulcerans_Agy99 RSIARGIAERYLRPGQIEEHLIYPVTTETTDHPGGYAGALEDFYEQATER
MAV_2430|M.avium_104 RSIARGIAEPYLRPGQIEEHLVYPVTTEVTDHPGGYAGAIEDFYVEATAR
MSMEG_3873|M.smegmatis_MC2_155 QSIARGIAEPYLRPGQLEEHLVYPVTTETTDHPGGYAGAMEDFYAEAAER
TH_0440|M.thermoresistible__bu RSIARSIAEPYLRPGQLEEHLVYPVTTETTDHPGGYAGAMEDFYTEAADR
Mflv_3104|M.gilvum_PYR-GCK RSIARSIAEQYLRDGQIEEHLVYPVTTETSEHPGGYAGAMEDFYRESADR
Mvan_3431|M.vanbaalenii_PYR-1 RSIARSIAEPYLREGQIEEHLVYPVTTETSDHPGGYAGAMEDFYRESADR
MAB_2198c|M.abscessus_ATCC_199 RSIARSIAEPYLRPGQIEEHLVYPVTTETTDHPGGYRGAIDDFYEESANR
:****.*** *** **:****:******.::***** **::*** ::: *
Rv2066|M.tuberculosis_H37Rv IATHLDAGRNVALLAEGDPLFYSSYMHLHTRLTRRFNAVIVPGVTSVSAA
Mb2092|M.bovis_AF2122/97 IATHLDAGRNVALLAEGDPLFYSSYMHLHTRLTRRFNAVIVPGVTSVSAA
MMAR_3046|M.marinum_M IATHLDAGRNVALLAEGDPLFYSSYMHLHTRLTQRFDAVIVPGVTSVSAA
MUL_2290|M.ulcerans_Agy99 IAAHLDAGRNVALLAEGDPLFYSSYVHLHTRLTQRFDAVIVPGVTSVSAA
MAV_2430|M.avium_104 IAAHLDAGRDVALLAEGDPLFYSSYMHLHTRLTQRFDAVIVPGVTSVSAA
MSMEG_3873|M.smegmatis_MC2_155 IAAHLQAGRDVALLAEGDPLFYSSYMHMHTRLTERFDAVIVPGVTSVSAA
TH_0440|M.thermoresistible__bu IAAHLEAGRDVALLAEGDPLFYSSYMHMHTRLTERFDAVIVPGVTSVSAA
Mflv_3104|M.gilvum_PYR-GCK IAAHLDAGRDVALLAEGDPLFYSSYMHMHTRLTERFDAIIVPGVTSVSAA
Mvan_3431|M.vanbaalenii_PYR-1 IAAHLDAGRDVALLAEGDPLFYSSYMHMHTRLTERFDALIVPGVTSVSAA
MAB_2198c|M.abscessus_ATCC_199 IAAHLDSGRNVALLAEGDPLFYSSYMHMHRRLTDRFDAVIIPGVTSFSAA
**:**::**:***************:*:* *** **:*:*:*****.***
Rv2066|M.tuberculosis_H37Rv SAAVATPLVAGDQVLSVLPGTLPVGELTRRLADADAAVVVKLGRSYHNVR
Mb2092|M.bovis_AF2122/97 SAAVATPLVAGDQVLSVLPGTLPVGELTRRLADADAAVVVKLGRSYHNVR
MMAR_3046|M.marinum_M SAAIATPLVAGDEVLSVLPGTLPVAELTRRLADVDAAVVLKLGRSYHNVR
MUL_2290|M.ulcerans_Agy99 SAAIATPLVAGDEVLSVLPGTLPVAELTRRLADVDAAVVLKLGRSYHNVR
MAV_2430|M.avium_104 SAAIATPLVAGDEVLSVLPGTLPVTELTRRLADADAAVVLKLGRSYHAVR
MSMEG_3873|M.smegmatis_MC2_155 SAATGTPLVQGDEVLTILPGTLPADELKRRLADTDAAVVMKLGRSYTRVR
TH_0440|M.thermoresistible__bu SAATGTPLVQGDEVLSILPGTLPADELTRRLADTDAAVVLKLGRSYPTVR
Mflv_3104|M.gilvum_PYR-GCK SAAVSTPLVQGDQVLTILPGTLPQAELRRRLADTDAAVILKLGRSYPAVR
Mvan_3431|M.vanbaalenii_PYR-1 SAAVSTPLVQGDQVLTILPGTLPADELRRRLADTDAAVILKLGRSYPAVR
MAB_2198c|M.abscessus_ATCC_199 SAATGTPLVEGDEILTVIPGTLPADEITRRLGDSHAAVIMKLGRSYGAVR
*** .**** **::*:::***** *: ***.* .***::****** **
Rv2066|M.tuberculosis_H37Rv EALSASGLLGDAFYVERASTAGQRVLPAADVDETSVPYFSLAMLPGGR--
Mb2092|M.bovis_AF2122/97 EALSASGLLGDAFYVERASTAGQRVLPAADVDETSVPYFSLAMLPGGR--
MMAR_3046|M.marinum_M EALSASGQLDDTFYVERASTPGQRVLPAADVDEASVPYFSLAMLPGGR--
MUL_2290|M.ulcerans_Agy99 EALSTSGQLDDTFYVERASTPGQRVLPAADVDEASVPYFSLAMLPGGR--
MAV_2430|M.avium_104 EALSQSGQLDDAVYVERASTSAQRIVPAADVDADGVPYFSLAILPGGQRA
MSMEG_3873|M.smegmatis_MC2_155 EALSSAGRLDEAFYVERASTDRQKVAQAGEVESAKVPYFSLTMVPGGM--
TH_0440|M.thermoresistible__bu QALSDAGRLDEAFYVERASTPAQRVAPAGEVDAERVPYFSLAMVPSAV--
Mflv_3104|M.gilvum_PYR-GCK QALSEAGRLDDAFYVERASTERQRVLPAADVDADSVPYFALAMLTGDG--
Mvan_3431|M.vanbaalenii_PYR-1 QALSAAGRIDDAFYVERASTERQRVLPASEVDDQSVPYFALTMLPGGR--
MAB_2198c|M.abscessus_ATCC_199 QALDASGLLDRAYYVERASTPGQRVLPAAEVDPASVPYFSLVLVPGAQS-
:**. :* :. : ******* *:: *.:*: ****:*.::..
Rv2066|M.tuberculosis_H37Rv RRALL---TGTVAVVGLGPGDSDWMTPQSRRELAAATDLIGYRGYLDRVE
Mb2092|M.bovis_AF2122/97 RRALL---TGTVAVVGLGPGDSDWMTPQSRRELAAATDLIGYRGYLDRVE
MMAR_3046|M.marinum_M RSTVV---AGTVAVVGLGPGDTDWMTPQSRRELARATDLIGYGPYLDRIS
MUL_2290|M.ulcerans_Agy99 RSTVV---AGTVAVVGLGPGDTDWMTPQSRRELARATDLIGYGPYLDRIS
MAV_2430|M.avium_104 RRAPG---AGTVAVVGLGPGDAQWMTPQSRRELAAATDLVGYGRYLDRVP
MSMEG_3873|M.smegmatis_MC2_155 ARQEQRPERGSVVVVGLGPGDHDWMTPQSRRELAAATDLIGYGPYLDRVG
TH_0440|M.thermoresistible__bu NRP-ARTGTGSVAVVGLGPGDPDWMTPQSRRELAAATDLIGYGPYLDRIP
Mflv_3104|M.gilvum_PYR-GCK RRR---VGTGSVAVVGLGPGDTAWMTPQSRSELAAATDLIGYGPYLNRVG
Mvan_3431|M.vanbaalenii_PYR-1 RAA---SVTGSVAVVGLGPGDAGWTTPQSRQVLAAATDLIGYGPYLDRVP
MAB_2198c|M.abscessus_ATCC_199 ATDIDTDPDGMVTVLGLGPGHHDWTTTEVRQELARATDLIGYGPYLDRIP
* *.*:*****. * *.: * ** ****:** **:*:
Rv2066|M.tuberculosis_H37Rv VRDGQRRHPSDNTDEPARARLACSLADQGRAVAVVSSGDPGVFAMATAVL
Mb2092|M.bovis_AF2122/97 VRDGQRRHPSDNTDEPARARLACSLADQGRAVAVVSSGDPGVFAMATAVL
MMAR_3046|M.marinum_M VRDGQRRHPSDNTDEPARARLACSLAEQGRAVAVVSSGDPGVFAMATAVL
MUL_2290|M.ulcerans_Agy99 VRDGQRRHPSDNTDEPARARLACSLAEQGRAVAVVSSGDPGVFAMATAVL
MAV_2430|M.avium_104 VRDGQRRHPSDNRDEPERARLACALAEQGRAVAVVSSGDPGVFAMATAVL
MSMEG_3873|M.smegmatis_MC2_155 LRPGQRHHPSDNTDEPERARLACTLAQQGHAVAVVSSGDPGVFAMATAVL
TH_0440|M.thermoresistible__bu VRAGQRRHPSDNTDEPARARLACSLAEQGCAVAVVSSGDPGVFAMATAVL
Mflv_3104|M.gilvum_PYR-GCK VREGQRRHPSDNTDEPARARLACELAEQGRAVAVVSSGDPGVFAMATAVL
Mvan_3431|M.vanbaalenii_PYR-1 VRDGQRRHPSDNTDEPARARLACELAEQGRTVAVVSSGDPGVFAMATAVL
MAB_2198c|M.abscessus_ATCC_199 ARDGQVRHASDNRDEPARAELAFKLAAGGRRVAVVSSGDPGVFAMAAAVL
* ** :*.*** *** **.** ** * ***************:***
Rv2066|M.tuberculosis_H37Rv EEAEQWPGVRVRVIPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWDVI
Mb2092|M.bovis_AF2122/97 EEAEQWPGVRVRVIPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWDVI
MMAR_3046|M.marinum_M EEAKQWPGVPVRVIPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWEVI
MUL_2290|M.ulcerans_Agy99 EEAKQWPGVPVRVIPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWEVI
MAV_2430|M.avium_104 EEAKQWPGVQVRVIPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWEVI
MSMEG_3873|M.smegmatis_MC2_155 EEAKQWPGVEVRVIPAMTAAQAVASRAGAPLGHDYAVISLSDRLKPWDVI
TH_0440|M.thermoresistible__bu EEARAWPGVQVRVVPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWDVI
Mflv_3104|M.gilvum_PYR-GCK EEAEQWPGVDVRVIPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWDVI
Mvan_3431|M.vanbaalenii_PYR-1 EEAEQWPDVDVRVVPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWDII
MAB_2198c|M.abscessus_ATCC_199 EEARAWPDVTVRVLPAMTAAQAVASRVGAPLGHDYAVISLSDRLKPWKVI
***. **.* ***:************.********************.:*
Rv2066|M.tuberculosis_H37Rv AARLTAAAAADLVLAIYNPASVTRTWQVGAMRELLLAHRDPGIPVVIGRN
Mb2092|M.bovis_AF2122/97 AARLTAAAAADLVLAIYNPASVTRTWQVGAMRELLLAHRDPGIPVVIGRN
MMAR_3046|M.marinum_M AARLGAAAAADLVLAIYNPASKTRTWQVGAMRELLLQHRDPGTPVVIGHN
MUL_2290|M.ulcerans_Agy99 AARLGAAAAADLVLAIYNPASKTRAWQVGAMRELLLQHRDPGTPVVIGRN
MAV_2430|M.avium_104 AARLEAAAAADLVLAIYNPASKARTWQVGAMRDILLARREPGTPVVIGRD
MSMEG_3873|M.smegmatis_MC2_155 VERLKAAAKADLVLAIYNPASKTRTWQVGAMRDLLLEHRDPSTPVIIGRA
TH_0440|M.thermoresistible__bu AARLKAAASADLVLAIYNPASKTRTWQVGAMRDLLLEHRDPATPVVIGRD
Mflv_3104|M.gilvum_PYR-GCK ARRLAAAAEADMVLAIYNPASKTRTWQVGAMRDLLLEHRDPATPVVIGRD
Mvan_3431|M.vanbaalenii_PYR-1 ARRLTAAAAADMVLAVYNPASKTRNWQVAAMRDLLLEHRDADTPVVIGRD
MAB_2198c|M.abscessus_ATCC_199 ESRLRAAAAADLVLAVYNPASKSRTWQVASMRDVLLEHRDPATPVVLGRD
** *** **:***:***** :* ***.:**::** :*:. **::*:
Rv2066|M.tuberculosis_H37Rv VSGPVSGPNEDVRVVKLADLNPAEIDMRCLLIVGSSQTRWYSVD----SQ
Mb2092|M.bovis_AF2122/97 VSGPVSGPNEDVRVVKLADLNPAEIDMRCLLIVGSSQTRWYSVD----SQ
MMAR_3046|M.marinum_M VS----GPDEAVRVVRLADLNPADVDMRCLLIIGSSQTQWYAVD----ST
MUL_2290|M.ulcerans_Agy99 VS----GPDEDVRVVRLADLNPADVDMRCLLIIGSSQTQWYAVD----ST
MAV_2430|M.avium_104 VS----GPDEDVRVVRLADLDPAEIDMRCLLIVGSSQTQWYSDD----SA
MSMEG_3873|M.smegmatis_MC2_155 VSGARPGEGESVRVVRLAELDPAQVDMRCLLIIGSSQTQWYSAAVDDTAE
TH_0440|M.thermoresistible__bu VSGPQ----ESVRVVRLAELDPADVDMRCLLIIGSSQTRWYTD-----GG
Mflv_3104|M.gilvum_PYR-GCK VAGPR----ESVTVVALGDLDPATVDMRCLLIIGSTQTRWQRTT---DGA
Mvan_3431|M.vanbaalenii_PYR-1 VAGPR----ESVRVVPLGELDPADVDMRCLLIVGSSRTRWHTTT---DGA
MAB_2198c|M.abscessus_ATCC_199 VGGAG----ESVRVTTLGELDPTVVDMRTLLIIGSSQTQWHDTD----SG
*. * * *. *.:*:*: :*** ***:**::*:* .
Rv2066|M.tuberculosis_H37Rv DRVFTPRRYPEAGRATATKSSRHSD
Mb2092|M.bovis_AF2122/97 DRVFTPRRYPEAGRATATKSSRHSD
MMAR_3046|M.marinum_M DRVFTPRRYP---------------
MUL_2290|M.ulcerans_Agy99 DRVFTPRHYP---------------
MAV_2430|M.avium_104 DRVFTPRRYEM--------------
MSMEG_3873|M.smegmatis_MC2_155 DRVFTPRTYPG--------------
TH_0440|M.thermoresistible__bu DRVFTPRRYPG--------------
Mflv_3104|M.gilvum_PYR-GCK DRVFTLRRYPT--------------
Mvan_3431|M.vanbaalenii_PYR-1 DRVFTPRTYPG--------------
MAB_2198c|M.abscessus_ATCC_199 PQVFTPRRYPAR-------------
:*** * *