For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TYVIGSECVDVMDKSCVQECPVDCIYEGARMLYINPDECVDCGACKPACRVEAIYWEGDLPDDQHQHLGD NAAFFHQVLPGRVAPLGSPGGAAAVGPIGVDTPLVAAIPVECP*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
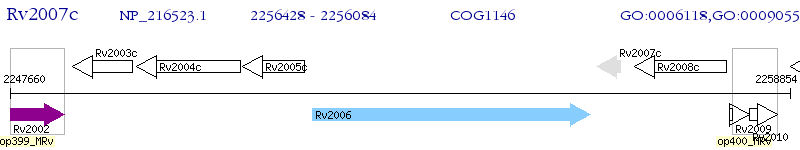
| M. tuberculosis H37Rv | Rv2007c | fdxA | - | 100% (114) | PROBABLE FERREDOXIN FDXA |
| M. tuberculosis H37Rv | Rv1177 | fdxC | 2e-32 | 61.86% (97) | ferredoxin FdxC |
| M. tuberculosis H37Rv | Rv0886 | fprB | 5e-10 | 40.24% (82) | NADPH:adrenodoxin oxidoreductase FprB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2030c | fdxA | 4e-68 | 100.00% (114) | ferredoxin FDXA |
| M. gilvum PYR-GCK | Mflv_0383 | - | 2e-44 | 70.00% (110) | 4Fe-4S ferredoxin iron-sulfur binding domain-containing protein |
| M. leprae Br4923 | MLBr_01489 | fdxA | 1e-32 | 61.86% (97) | ferredoxin |
| M. abscessus ATCC 19977 | MAB_1327 | - | 5e-35 | 59.09% (110) | ferredoxin FdxC |
| M. marinum M | MMAR_2994 | fdxA_2 | 7e-51 | 78.18% (110) | ferredoxin FdxA_2 |
| M. avium 104 | MAV_3500 | - | 3e-47 | 72.73% (110) | putative ferredoxin FdxA |
| M. smegmatis MC2 155 | MSMEG_1124 | - | 8e-44 | 68.18% (110) | putative ferredoxin FdxA |
| M. thermoresistible (build 8) | TH_0923 | fdxA | 5e-48 | 73.64% (110) | PROBABLE FERREDOXIN FDXA |
| M. ulcerans Agy99 | MUL_3264 | fdxA_1 | 3e-44 | 68.18% (110) | ferredoxin FdxA_1 |
| M. vanbaalenii PYR-1 | Mvan_0299 | - | 8e-46 | 71.82% (110) | 4Fe-4S ferredoxin iron-sulfur binding domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0383|M.gilvum_PYR-GCK MQSRRDQTRDFRPLTTTAARPTVDGMTYVIGSACVDIVDKSCVQECPADC
Mvan_0299|M.vanbaalenii_PYR-1 -------------------------MTYVIGSACVDIVDKSCVQECPADC
TH_0923|M.thermoresistible__bu -------------------------MTYVIGSACVDVTDKSCVQECPADC
Rv2007c|M.tuberculosis_H37Rv -------------------------MTYVIGSECVDVMDKSCVQECPVDC
Mb2030c|M.bovis_AF2122/97 -------------------------MTYVIGSECVDVMDKSCVQECPVDC
MMAR_2994|M.marinum_M -------------------------MTYVIGRECVDVAEKSCTQECPVDC
MAV_3500|M.avium_104 -------------------------MTYVIGKPCIDVMDRACVDECPVDC
MUL_3264|M.ulcerans_Agy99 -------------------------MTYVIGKPCIDVTDRACVEECPVDC
MLBr_01489|M.leprae_Br4923 -------------------------MTYVIAEPCVDIKDKACIEECPVDC
MAB_1327|M.abscessus_ATCC_1997 ----------MYWTGNSPENIEETFVTYTIAEPCVDVMDKACIEECPVDC
MSMEG_1124|M.smegmatis_MC2_155 -------------------------MTYVIGRPCVDVKDRACVDECPVDC
:**.*. *:*: :::* :***.**
Mflv_0383|M.gilvum_PYR-GCK IYEGDRAMYINPNECVDCGACRIACRVDAIYYETDLPDEELAFLDDNAAF
Mvan_0299|M.vanbaalenii_PYR-1 IYEGDRAMYINPNECVDCGACRIACRVDAIYYETDLPDEEMEFLEDNAAF
TH_0923|M.thermoresistible__bu IYEGDRSLYINPNECVDCGACKLVCKVDAIYFEYDLPDEEYRHLADNAAF
Rv2007c|M.tuberculosis_H37Rv IYEGARMLYINPDECVDCGACKPACRVEAIYWEGDLPDDQHQHLGDNAAF
Mb2030c|M.bovis_AF2122/97 IYEGARMLYINPDECVDCGACKPACRVEAIYWEGDLPDDQHQHLGDNAAF
MMAR_2994|M.marinum_M IYEGARTMYINPDECVDCGACKTTCRVGAIYWEEDLPDEQRHHLADNAAF
MAV_3500|M.avium_104 IYEGGRALYIHPDECVDCGACEPVCPVEAIYYEDDLPEDLKPHLADNEAF
MUL_3264|M.ulcerans_Agy99 IYEGGRSLYIHPDEFVDCGACEPVCPVEAIYYEDDLPQELHPHLADNVAF
MLBr_01489|M.leprae_Br4923 IYEGARMLYIHPDECVDCGACEPVCPVEAIYYEDDVPEQWSHYTQINVDF
MAB_1327|M.abscessus_ATCC_1997 IYEGGRMLYIHPDECVDCGACEPVCPVEAIFYEDDVPDQWTGYIQSNADF
MSMEG_1124|M.smegmatis_MC2_155 IYEGARMLYIHPDECVDCGACEPVCPVEAIYYEDDLPEDLQPYQEENAKF
**** * :**:*:* ******. .* * **::* *:*:: . * *
Mflv_0383|M.gilvum_PYR-GCK FTTTLSGRDEPLGDPGGAAKLGRVGADTPLVAALPASTTPHP-
Mvan_0299|M.vanbaalenii_PYR-1 FTMTLAGRDAPLGDPGGAAKVGRVGADTPLVAALPPSTTPHP-
TH_0923|M.thermoresistible__bu FTEILPGRDAPLGDPGGASKIGRIGVDTPLVAGMPK-RSAH--
Rv2007c|M.tuberculosis_H37Rv FHQVLPGRVAPLGSPGGAAAVGPIGVDTPLVAAIPVECP----
Mb2030c|M.bovis_AF2122/97 FHQVLPGRVAPLGSPGGAAAVGPIGVDTPLVAAIPVECP----
MMAR_2994|M.marinum_M FREILPGRDAPLGSPGGADTVGRIGVDTPLIAAMPPSGL----
MAV_3500|M.avium_104 FAEPLPGRDAPLGSPGGAAKLGPLGVDTPLVASQPSAAHSDGS
MUL_3264|M.ulcerans_Agy99 FTETLPGRDGPLGSPGGAAKISRLGVDTPLVAGHPQAADA---
MLBr_01489|M.leprae_Br4923 FVE--------LGSPGGAAKVGMAENDPQVIKNLAPQGEGD--
MAB_1327|M.abscessus_ATCC_1997 FVD--------LGSPGGAAKVGKTDYDPPSVKELPPMGEGH--
MSMEG_1124|M.smegmatis_MC2_155 FTDVLPGRAQPLGSPGGAAKLGVVDADTPMVAELPPQGD----
* **.**** :. *. : .