For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNSPLVVGFLACFTLIAAIGAQNAFVLRQGIQREHVLPVVALCTVSDIVLIAAGIAGFGALIGAHPRALN VVKFGGAAFLIGYGLLAARRAWRPVALIPSGATPVRLAEVLVTCAAFTFLNPHVYLDTVVLLGALANEHS DQRWLFGLGAVTASAVWFATLGFGAGRLRGLFTNPGSWRILDGLIAVMMVALGISLTVT
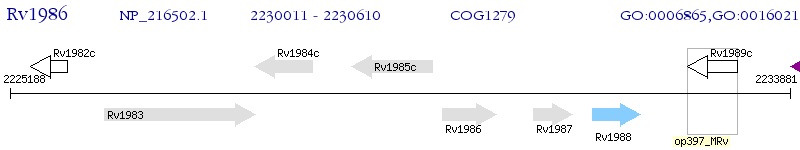
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1986 | - | - | 100% (199) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. tuberculosis H37Rv | Rv0488 | - | 6e-64 | 63.44% (186) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2008 | - | 1e-110 | 100.00% (199) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_0373 | - | 2e-74 | 71.96% (189) | lysine exporter protein LysE/YggA |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4386 | - | 4e-69 | 65.64% (195) | putative transporter |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0467 | - | 9e-80 | 73.37% (199) | membrane transport protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0310 | - | 2e-76 | 70.30% (202) | lysine exporter protein LysE/YggA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0373|M.gilvum_PYR-GCK MSTTSLVVLSGFASSMALIAAIGAQNAFVLRQGIRGEHVLPVVAVCAVSD
Mvan_0310|M.vanbaalenii_PYR-1 MNPAYVVVLSGFATTMALIAAIGAQNAFVLRQGIRGEHVLPVVAVCTVSD
Rv1986|M.tuberculosis_H37Rv MNSP---LVVGFLACFTLIAAIGAQNAFVLRQGIQREHVLPVVALCTVSD
Mb2008|M.bovis_AF2122/97 MNSP---LVVGFLACFTLIAAIGAQNAFVLRQGIQREHVLPVVALCTVSD
MSMEG_0467|M.smegmatis_MC2_155 MNSP---LVLGFLTSMALIAAIGAQNAFVLRQGIRREHVLPVIAVCTVSD
MAB_4386|M.abscessus_ATCC_1997 MTVL----FLGFLTGMSLIAAIGAQNAFVLRQGIRSEHVLPVIAVCITGD
*. . ** : ::*****************: ******:*:* ..*
Mflv_0373|M.gilvum_PYR-GCK LVLIAAGIAGVGALIAAHPDVVTVTRYGGAAFLIGYGLFAARRAVKPGVL
Mvan_0310|M.vanbaalenii_PYR-1 LVLISAGIAGFGALIAAHPDIVTATKFGGAAFLVGYGLLAARRAVKPGVL
Rv1986|M.tuberculosis_H37Rv IVLIAAGIAGFGALIGAHPRALNVVKFGGAAFLIGYGLLAARRAWRPVAL
Mb2008|M.bovis_AF2122/97 IVLIAAGIAGFGALIGAHPRALNVVKFGGAAFLIGYGLLAARRAWRPVAL
MSMEG_0467|M.smegmatis_MC2_155 LLLITAGIAGVGAVITAHPDAVTVTKFGGAAFLIGYGVLAARRALRPSTL
MAB_4386|M.abscessus_ATCC_1997 FLLIAAGIGGLGGLITATPSLLTVAKVGGAAFLIGYGLMAARRALRPGTL
::**:***.*.*.:* * * :...: ******:***::***** :* .*
Mflv_0373|M.gilvum_PYR-GCK TPADAAPARLAAVLATCLALTFLNPHVYLDTVVLLGMLANEHQDSRWLFG
Mvan_0310|M.vanbaalenii_PYR-1 TPADAAPARLTAVLATCLAMTFLNPHVYLDTVILLGALANEHQDSKWLFG
Rv1986|M.tuberculosis_H37Rv IPSGATPVRLAEVLVTCAAFTFLNPHVYLDTVVLLGALANEHSDQRWLFG
Mb2008|M.bovis_AF2122/97 IPSGATPVRLAEVLVTCAAFTFLNPHVYLDTVVLLGALANEHSDQRWLFG
MSMEG_0467|M.smegmatis_MC2_155 NPSERTPARLAEVLVTCLALTWLNPHVYLDTVVLLGTLANEQREQRWLFG
MAB_4386|M.abscessus_ATCC_1997 VPAESNPARLGAVLLTALAITFLNPHVYLDTVVLLGSLANAHRDARWLFG
*: *.** ** *. *:*:**********:*** *** : : :****
Mflv_0373|M.gilvum_PYR-GCK VGAVTASAVWFAGLGFGATRVRHWFASPRTWRLLDAAIAAVMIALGVSLL
Mvan_0310|M.vanbaalenii_PYR-1 VGAVAASALWFTGLGFGAKRLRHLFASPKTWRLLDGVIAATMLALGISLV
Rv1986|M.tuberculosis_H37Rv LGAVTASAVWFATLGFGAGRLRGLFTNPGSWRILDGLIAVMMVALGISLT
Mb2008|M.bovis_AF2122/97 LGAVTASAVWFATLGFGAGRLRGLFTNPGSWRILDGLIAVMMVALGISLT
MSMEG_0467|M.smegmatis_MC2_155 AGAVLASAIWFLGLGLGAKRLAGLFATPMTWRILDGVIAVTMIALGLGMM
MAB_4386|M.abscessus_ATCC_1997 AGALTASVTWFVGLGYGARYLAGVFERPGTWRMLDAGIAVVMLALGVSLL
**: **. ** ** ** : * * :**:**. **. *:***:.:
Mflv_0373|M.gilvum_PYR-GCK MR
Mvan_0310|M.vanbaalenii_PYR-1 VS
Rv1986|M.tuberculosis_H37Rv VT
Mb2008|M.bovis_AF2122/97 VT
MSMEG_0467|M.smegmatis_MC2_155 LT
MAB_4386|M.abscessus_ATCC_1997 VS
: