For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKTTDVRVRRAITAMAGGHAVVLTGDPNGDGYLVFAAQAATPRLVAFAVRHTSGYLRVALPGAECERLHL PPMCDRDTTHCVSVDVRGTGTGISASDRAWTIAALASATSVAADFQRPGHVVPVQAQADGVLGRRGPAEA AVDLARLAERRPAAALCEIVSPDNPVQMAHHAESVEFAVEHGLAMVSIGELVAYRRRIEPQVVRFTAATL PTWAGASRVIGFRDVYDLGEHLAVIVGAVGAGVPVPLHVHIECLTGDVFGSTACRCGEELNGALARMSAQ GSGVVLYLRPPGPAQACGLFARGDAATDVMPETVTWILRDLGVYAIRLSDDVPGFGLVMFGAIREASTLA AAG
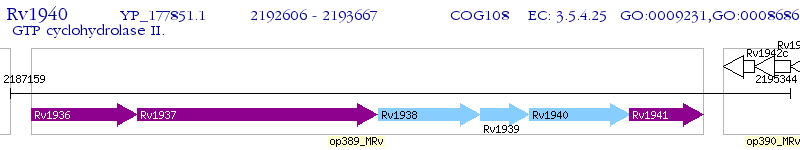
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1940 | ribA1 | - | 100% (353) | Probable Riboflavin biosynthesis protein ribA1 (GTP cyclohydrolase II) |
| M. tuberculosis H37Rv | Rv1415 | ribA2 | 1e-70 | 44.57% (368) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1975 | ribA1 | 0.0 | 100.00% (353) | riboflavin biosynthesis protein ribA1 (GTP cyclohydrolase |
| M. gilvum PYR-GCK | Mflv_3719 | - | 4e-67 | 44.29% (368) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. leprae Br4923 | MLBr_00559 | ribA | 1e-68 | 44.02% (368) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate |
| M. abscessus ATCC 19977 | MAB_2796c | - | 2e-68 | 48.64% (294) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. marinum M | MMAR_2868 | ribA1 | 1e-143 | 74.79% (357) | riboflavin biosynthesis protein RibA1 |
| M. avium 104 | MAV_3365 | - | 4e-69 | 48.98% (294) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. smegmatis MC2 155 | MSMEG_3072 | - | 2e-67 | 48.98% (294) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. thermoresistible (build 8) | TH_1011 | - | 9e-68 | 49.32% (294) | riboflavin biosynthesis protein ribAB |
| M. ulcerans Agy99 | MUL_2886 | ribA1 | 1e-143 | 74.23% (357) | riboflavin biosynthesis protein RibA1 |
| M. vanbaalenii PYR-1 | Mvan_4000 | - | 1e-123 | 64.17% (360) | 3,4-dihydroxy-2-butanone 4-phosphate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00559|M.leprae_Br4923 MGPFDSKVAKMTRLDS-VERAVADIAAGKAVVVIDDEDRENEGDLIFAAE
MAV_3365|M.avium_104 ----------MTRLDS-VERAVADIAAGKAVIVIDDEDRENEGDLIFAAE
Mflv_3719|M.gilvum_PYR-GCK ----------MTRLDS-VERAVADIAAGKAVVVIDDEDRENEGDLIFAAE
MSMEG_3072|M.smegmatis_MC2_155 ----------MTRLDS-VERAIADIAAGKAVVVIDDEDRENEGDLIFAAE
TH_1011|M.thermoresistible__bu ----------MVRLDS-VDRAVADIAAGKAVVVIDDEDRENEGDLIFAAE
MAB_2796c|M.abscessus_ATCC_199 ----------MTRLDS-IERAVADIAAGKAVVVVDDEDRENEGDLIFAAE
Rv1940|M.tuberculosis_H37Rv ----------MKTTDVRVRRAITAMAGGHAVVLTGDPN--GDGYLVFAAQ
Mb1975|M.bovis_AF2122/97 ----------MKTTDVRVRRAITAMAGGHAVVLTGDPN--GDGYLVFAAQ
MMAR_2868|M.marinum_M ----------MKTTDVRVRRAITAIAAGRPVVVTHDHA--AQGYLVFAAE
MUL_2886|M.ulcerans_Agy99 ----------MKTTDVRVRRAITAIAAGRPVVVTHDHA--AQGYLVFAAE
Mvan_4000|M.vanbaalenii_PYR-1 ----------MRTTDLRVRRAIRAVAEGRPVVVTADTEREGAGYLVFAAD
* * : **: :* *:.*:: * * *:***:
MLBr_00559|M.leprae_Br4923 KATLEMVAFMVRYTSGYLCAPLDGAICDRLGLLPMFAVNQD-KHGTAYTV
MAV_3365|M.avium_104 KATPEMVAFMVRYTSGYLCVPLDGAICDRLGLLPMYAVNQD-KHGTAYTV
Mflv_3719|M.gilvum_PYR-GCK KATPELVAFMVRYTSGYLCVPLAGEICDRLGLLPMYAVNQD-KHGTAYTV
MSMEG_3072|M.smegmatis_MC2_155 KATPELVAFMVRYTSGYLCVPLDGEICDRLGLLPMYAVNQD-KHGTAYTV
TH_1011|M.thermoresistible__bu KATPELVAFMVRYTSGYLCVPLEGEICDRLGLLPMYAMNQD-KHGTAYTV
MAB_2796c|M.abscessus_ATCC_199 KATPELVAFMVRYTSGYLCVPLAGEDCDRLGLLPQYAVNQD-KHGTAYTV
Rv1940|M.tuberculosis_H37Rv AATPRLVAFAVRHTSGYLRVALPGAECERLHLPPMCDRDT------THCV
Mb1975|M.bovis_AF2122/97 AATPRLVAFAVRHTSGYLRVALPGAECERLHLPPMCDRDT------THCV
MMAR_2868|M.marinum_M AATSSLVAFTVRHTAGYVRVALPGAECARLNLPPMCHHNP------THRV
MUL_2886|M.ulcerans_Agy99 AATSSLVAFTVRHTAGYVRVALPGAECARLNLPPMCHHNP------THRV
Mvan_4000|M.vanbaalenii_PYR-1 AATPNILAFTVRHTSGLVRVALPLSECARLDLPPMCSADPDGSHAAAQRV
** ::** **:*:* : ..* * ** * * : : *
MLBr_00559|M.leprae_Br4923 TVDARNGVGTGISASDRATTMRLLADPASVADDFTRPGHVVPLRAKDGGV
MAV_3365|M.avium_104 TVDARNGVGTGISASDRATTMRLLADPTSIAEDFTRPGHVVPLRAKDGGV
Mflv_3719|M.gilvum_PYR-GCK TVDAKVGVGTGISASDRATTMRLLADPDSVADDFTKPGHVVPLRAKDGGV
MSMEG_3072|M.smegmatis_MC2_155 TVDAKKDVGTGISASDRATTMRALANPGSVADDFTKPGHVVPLRAKDGGV
TH_1011|M.thermoresistible__bu TVDAKHGVTTGISAADRATTMRLLADPNAVADDFTRPGHVVPLRAKEGGV
MAB_2796c|M.abscessus_ATCC_199 TVDAKNGIGTGISASDRAATMRLLADAASTADDFTKPGHVVPLRAKDGGV
Rv1940|M.tuberculosis_H37Rv SVDVR-GTGTGISASDRAWTIAALASATSVAADFQRPGHVVPVQAQADGV
Mb1975|M.bovis_AF2122/97 SVDVR-GTGTGISASDRAWTIAALASATSVAADFQRPGHVVPVQAQADGV
MMAR_2868|M.marinum_M SVDVR-ATGTGISARDRARTIAALASAGSQAADFLRPGHVAPLLAEPHGV
MUL_2886|M.ulcerans_Agy99 SVDVR-ATGTGISARDRARTIAALASAGSQAADFLRPGHVAPLLAEPHGV
Mvan_4000|M.vanbaalenii_PYR-1 AVDAG-GNGTGISATDRARTIAALASEHAQPGDFRRPGHVIPVQANADGV
:**. ***** *** *: **. : . ** :**** *: *: **
MLBr_00559|M.leprae_Br4923 LRRPGHTEAAVDLARLAGLQPAGAICEIVSQKDEGSMAQTDELRVFADEH
MAV_3365|M.avium_104 LRRPGHTEAAVDLARMAGLQPAGAICEIVSQKDEGSMAQTDELRVFADEH
Mflv_3719|M.gilvum_PYR-GCK LRRPGHTEAAVDLARLAGLQAAGTICEIVSQKDEGAMAQTDELRIFADEH
MSMEG_3072|M.smegmatis_MC2_155 LRRPGHTEAAVDLARLAGLQPAGAICEIVSQKDEGDMARTDELRVFADDH
TH_1011|M.thermoresistible__bu LRRPGHTEAAVDLAKLAGLQPAGAICEIVSQKDEGAMAKTDELRVFADDH
MAB_2796c|M.abscessus_ATCC_199 LRRPGHTEASVDLAKLAGLRPAAVICEIVSQKDEGAMAQTEELRVFADEH
Rv1940|M.tuberculosis_H37Rv LGRRGPAEAAVDLARLAERRPAAALCEIVSPDNPVQMAHHAESVEFAVEH
Mb1975|M.bovis_AF2122/97 LGRRGPAEAAVDLARLAERRPAAALCEIVSPDNPVQMAHHAESVEFAVEH
MMAR_2868|M.marinum_M LAKCEPAEAAVDLARLAGHRPAAALCEIVSRRNPTLMAHGAELVEFAVEH
MUL_2886|M.ulcerans_Agy99 LAKCEPAEAAVDLARLAGHRPAAALCEIVSRRNPTLMAHGAELVEFAVEH
Mvan_4000|M.vanbaalenii_PYR-1 LGSPGAVEAAVDLARLSGRRQAAGLCEIVSRNHPATIACGTEPVEFALEH
* .**:****::: : *. :***** . :* * ** :*
MLBr_00559|M.leprae_Br4923 DLALITIADLIAWRRKHEKHIERVAEARIPTRHGEFRAIGYTSI---YDD
MAV_3365|M.avium_104 DLAMITIADLIEWRRKHEKHIERIAEARIPTRHGEFRAIGYTSI---YEE
Mflv_3719|M.gilvum_PYR-GCK DLALISIADLIEWRRKHEKHIARVAEARIPTRHGEFRAVGYTSI---YED
MSMEG_3072|M.smegmatis_MC2_155 DLALISIADLIEWRRKHEKHIERIAEARIPTRHGEFRAVGYKSI---YED
TH_1011|M.thermoresistible__bu GLAMISIADLIEWRRKHEKHVERVAEARIPTRHGEFRAVGYTSI---YDD
MAB_2796c|M.abscessus_ATCC_199 NLALVSIADLIEWRRKHEKHVQRIAEARIPTQHGEFRAVGYTSI---YDD
Rv1940|M.tuberculosis_H37Rv GLAMVSIGELVAYRRRIEPQVVRFTAATLPTWAGASRVIGFRDV---YDL
Mb1975|M.bovis_AF2122/97 GLAMVSIGELVAYRRRIEPQVVRFTAATLPTWAGASRVIGFRDV---YDL
MMAR_2868|M.marinum_M GLAVVSIAELLAYRQRTEPQVIRLTETTLPTWAGNSRVIGFRDLGPANPG
MUL_2886|M.ulcerans_Agy99 GLAVVSIAELLAYRQRTEPQVIRLTETTLPTWAGNSRVIGFRDLGPANPG
Mvan_4000|M.vanbaalenii_PYR-1 GLATVSIGELVAYRRRTERQVVRLAETVLPTGCGTHRVIGFRDV---HGG
.** ::*.:*: :*:: * :: *.: : :** * *.:*: .:
MLBr_00559|M.leprae_Br4923 VKHVALVRGEIAGSSGDGDDILVRVHSECLTGDVFGSRRCDCGTQLDAAM
MAV_3365|M.avium_104 VEHVALVRGEIAGPNSDGDDVLVRVHSECLTGDVFGSRRCDCGPQLDAAM
Mflv_3719|M.gilvum_PYR-GCK VEHVALVKGDIAGPHGDGHDVLVRVHSECLTGDVFGSRRCDCGPQLDAAL
MSMEG_3072|M.smegmatis_MC2_155 VEHVALVRGEISGPGSDGDDVLVRVHSECLTGDVFGSRRCDCGPQLDAAM
TH_1011|M.thermoresistible__bu VEHVALVMGDIAGPHGDGHDVLVRVHSECLTGDVFGSRRCDCGPQLDAAL
MAB_2796c|M.abscessus_ATCC_199 VEHVALVLGDISGPDGDGNDVLVRVHSECLTGDVFGSRRCDCGPQLDAAM
Rv1940|M.tuberculosis_H37Rv GEHLAVIVGAV----GAGVPVPLHVHIECLTGDVFGSTACRCGEELNGAL
Mb1975|M.bovis_AF2122/97 GEHLAVIVGAV----GAGVPVPLHVHIECLTGDVFGSTACRCGEELNGAL
MMAR_2868|M.marinum_M GEHLAVIIGAP----DSGVPVALHVHIECLTGDVFGSTGCRCGGELNGAL
MUL_2886|M.ulcerans_Agy99 GEHLAVVIGAP----DSGVPVALHVHIECLTGDVFGSTGCRCGGELNGAL
Mvan_4000|M.vanbaalenii_PYR-1 AEHLVLVIGSV----GSGEPVPLHVHVECLTGDVFGSTVCHCGAELDSAV
:*:.:: * . * : ::** ********** * ** :*:.*:
MLBr_00559|M.leprae_Br4923 AMVAREGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGADTVDANIKLGFP
MAV_3365|M.avium_104 AMVAREGRGIVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGFP
Mflv_3719|M.gilvum_PYR-GCK AMVAREGRGIVLYMRGHEGRGIGLMHKLQAYQLQDAGDDTVDANLKLGLP
MSMEG_3072|M.smegmatis_MC2_155 AMVAREGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGLP
TH_1011|M.thermoresistible__bu SMVAREGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGLP
MAB_2796c|M.abscessus_ATCC_199 EMVAKEGRGIVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGLP
Rv1940|M.tuberculosis_H37Rv ARMSAQGSGVVLYLR-------------PPGPAQACGLFARG-DAATDVM
Mb1975|M.bovis_AF2122/97 ARMSAQGSGVVLYLR-------------PPGPAQACGLFARG-DAATDVM
MMAR_2868|M.marinum_M SAMSAQGSGVVLYLR-------------PPGPPRACGLFASDEETPADFM
MUL_2886|M.ulcerans_Agy99 SAMSAQGSGVVLYLQ-------------PPGPPRACGLFASDEETPADFM
Mvan_4000|M.vanbaalenii_PYR-1 ATMSAQGGGVIVYLR-------------PPGSLRACGLPRPDHSPSPDLL
:: :* *:::*:: . : .* . . ..
MLBr_00559|M.leprae_Br4923 SDARDYGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLQIIERVPLPVR
MAV_3365|M.avium_104 ADARDYGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVR
Mflv_3719|M.gilvum_PYR-GCK ADARDYGTGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVR
MSMEG_3072|M.smegmatis_MC2_155 ADARDYGIGAQILVDLGIRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVR
TH_1011|M.thermoresistible__bu ADARDYGIGAQILVDLGVRSMRLLTNNPAKRAGLDGYGLHIIERVPLPVR
MAB_2796c|M.abscessus_ATCC_199 ADARDYGLGAQILVDLGVKSMRLLTNNPAKRVGLDGYGLHIIERVPLPVR
Rv1940|M.tuberculosis_H37Rv PETVTW-----ILRDLGVYAIRLSDDVP-------GFGLVMFG----AIR
Mb1975|M.bovis_AF2122/97 PETVTW-----ILRDLGVYAIRLSDDVP-------GFGLVMFG----AIR
MMAR_2868|M.marinum_M SETVTW-----MLRDLGVYAIKLSDDMP-------GFGLVMFG----AIR
MUL_2886|M.ulcerans_Agy99 SETVTW-----MLRDLGVYAIKLSDDMP-------GFGLVMFG----AIR
Mvan_4000|M.vanbaalenii_PYR-1 SDTVAW-----ILRDLGVYSLRLADDAP-------GYGLVMFG----ALR
.:: : :* ***: :::* : * *:** :: .:*
MLBr_00559|M.leprae_Br4923 ANADNIRYLMTKRDKMGHDLGGLDDFHESN--------------YFPGEF
MAV_3365|M.avium_104 ANAENIRYLMTKRDKMGHDLAGLDDFHESV--------------HLPGEF
Mflv_3719|M.gilvum_PYR-GCK ANAENIRYLMTKRDRMGHDLVGLDDFDEA----------------VPGEF
MSMEG_3072|M.smegmatis_MC2_155 ANKENIRYLMTKRDRMGHDLVGLDDYDEAVSMDDYDEAVYLLGDRRPTES
TH_1011|M.thermoresistible__bu ATADNIRYLMTKRDRMGHDLVGLEDFDESVRMDDYDEGVYLLGDRRPTDL
MAB_2796c|M.abscessus_ATCC_199 ANSENIRYLRTKRDRMGHDLADLDDHPEAD--------------------
Rv1940|M.tuberculosis_H37Rv EA--------STLAAAG---------------------------------
Mb1975|M.bovis_AF2122/97 EA--------STLAAAG---------------------------------
MMAR_2868|M.marinum_M EAGAAAGPADTPLAAAG---------------------------------
MUL_2886|M.ulcerans_Agy99 EAGAAAGPADRPLAAAG---------------------------------
Mvan_4000|M.vanbaalenii_PYR-1 QN----------RTAA----------------------------------
MLBr_00559|M.leprae_Br4923 --GGAL
MAV_3365|M.avium_104 --GGAL
Mflv_3719|M.gilvum_PYR-GCK --GGAV
MSMEG_3072|M.smegmatis_MC2_155 --GGAQ
TH_1011|M.thermoresistible__bu DLGGAL
MAB_2796c|M.abscessus_ATCC_199 ---GA-
Rv1940|M.tuberculosis_H37Rv ------
Mb1975|M.bovis_AF2122/97 ------
MMAR_2868|M.marinum_M ------
MUL_2886|M.ulcerans_Agy99 ------
Mvan_4000|M.vanbaalenii_PYR-1 ------