For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRLDSVERAVADIAAGKAVIVIDDEDRENEGDLIFAAEKATPEMVAFMVRYTSGYLCVPLDGAICDRLG LLPMYAVNQDKHGTAYTVTVDARNGIGTGISASDRATTMRLLADPTSVADDFTRPGHVVPLRAKDGGVLR RPGHTEAAVDLARMAGLQPAGAICEIVSQKDEGSMAHTDELRVFADEHGLALITIADLIEWRRKHEKHIE RVAEARIPTRHGEFRAIGYTSIYEDVEHVALVRGEIAGPNADGDDVLVRVHSECLTGDVFGSRRCDCGPQ LDAALAMVAREGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGADTVDANLKLGLPADARDYGIGAQILVD LGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAENIRYLMTKRDKLGHDLAGLDDFHESVHLPGE FGGAL
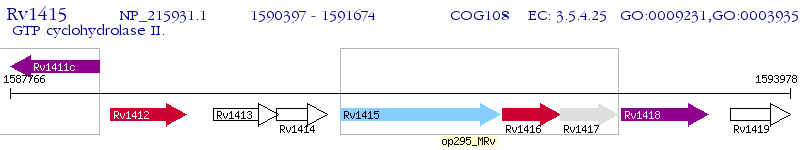
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1415 | ribA2 | - | 100% (425) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II protein |
| M. tuberculosis H37Rv | Rv1940 | ribA1 | 1e-70 | 44.57% (368) | riboflavin biosynthesis protein ribA1 (GTP cyclohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1450 | ribA2 | 0.0 | 100.00% (425) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate |
| M. gilvum PYR-GCK | Mflv_3719 | - | 0.0 | 91.76% (425) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. leprae Br4923 | MLBr_00559 | ribA | 0.0 | 93.41% (425) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate |
| M. abscessus ATCC 19977 | MAB_2796c | - | 0.0 | 87.20% (414) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. marinum M | MMAR_2222 | ribA2 | 0.0 | 96.47% (425) | riboflavin biosynthesis protein RibA2 |
| M. avium 104 | MAV_3365 | - | 0.0 | 96.71% (425) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. smegmatis MC2 155 | MSMEG_3072 | - | 0.0 | 91.37% (417) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
| M. thermoresistible (build 8) | TH_1011 | - | 0.0 | 89.93% (417) | riboflavin biosynthesis protein ribAB |
| M. ulcerans Agy99 | MUL_1809 | ribA2 | 0.0 | 95.29% (425) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate |
| M. vanbaalenii PYR-1 | Mvan_2694 | - | 0.0 | 91.53% (425) | bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3719|M.gilvum_PYR-GCK ----------MTRLDSVERAVADIAAGKAVVVIDDEDRENEGDLIFAAEK
Mvan_2694|M.vanbaalenii_PYR-1 ----------MTRLDSVERAVADIAAGKAVVVIDDEDRENEGDLIFAAEK
Rv1415|M.tuberculosis_H37Rv ----------MTRLDSVERAVADIAAGKAVIVIDDEDRENEGDLIFAAEK
Mb1450|M.bovis_AF2122/97 ----------MTRLDSVERAVADIAAGKAVIVIDDEDRENEGDLIFAAEK
MAV_3365|M.avium_104 ----------MTRLDSVERAVADIAAGKAVIVIDDEDRENEGDLIFAAEK
MLBr_00559|M.leprae_Br4923 MGPFDSKVAKMTRLDSVERAVADIAAGKAVVVIDDEDRENEGDLIFAAEK
MMAR_2222|M.marinum_M ----------MTRLDSVERAVADIGAGKAVIVIDDEDRENEGDLIFAAEK
MUL_1809|M.ulcerans_Agy99 ----------MTRLDSVERAVADIGAGKAVIVIDDEDRENEGDLIFAAEK
MSMEG_3072|M.smegmatis_MC2_155 ----------MTRLDSVERAIADIAAGKAVVVIDDEDRENEGDLIFAAEK
TH_1011|M.thermoresistible__bu ----------MVRLDSVDRAVADIAAGKAVVVIDDEDRENEGDLIFAAEK
MAB_2796c|M.abscessus_ATCC_199 ----------MTRLDSIERAVADIAAGKAVVVVDDEDRENEGDLIFAAEK
*.****::**:***.*****:*:*****************
Mflv_3719|M.gilvum_PYR-GCK ATPELVAFMVRYTSGYLCVPLAGEICDRLGLLPMYAVNQDKHGTAYTVTV
Mvan_2694|M.vanbaalenii_PYR-1 ATPELVAFMVRYTSGYLCVPLAGDICDRLGLLPMYAVNQDKHGTAYTVTV
Rv1415|M.tuberculosis_H37Rv ATPEMVAFMVRYTSGYLCVPLDGAICDRLGLLPMYAVNQDKHGTAYTVTV
Mb1450|M.bovis_AF2122/97 ATPEMVAFMVRYTSGYLCVPLDGAICDRLGLLPMYAVNQDKHGTAYTVTV
MAV_3365|M.avium_104 ATPEMVAFMVRYTSGYLCVPLDGAICDRLGLLPMYAVNQDKHGTAYTVTV
MLBr_00559|M.leprae_Br4923 ATLEMVAFMVRYTSGYLCAPLDGAICDRLGLLPMFAVNQDKHGTAYTVTV
MMAR_2222|M.marinum_M ATPELVAFMVRYTSGYLCVPLDGAVCDRLGLLPMYAVNQDKHGTAYTVTV
MUL_1809|M.ulcerans_Agy99 ATPELVAFMVRYTSGYLCVPLDGAVCDRLGLLPMYAVNQDKHGTAYTVTV
MSMEG_3072|M.smegmatis_MC2_155 ATPELVAFMVRYTSGYLCVPLDGEICDRLGLLPMYAVNQDKHGTAYTVTV
TH_1011|M.thermoresistible__bu ATPELVAFMVRYTSGYLCVPLEGEICDRLGLLPMYAMNQDKHGTAYTVTV
MAB_2796c|M.abscessus_ATCC_199 ATPELVAFMVRYTSGYLCVPLAGEDCDRLGLLPQYAVNQDKHGTAYTVTV
** *:*************.** * ******** :*:*************
Mflv_3719|M.gilvum_PYR-GCK DAKVGVGTGISASDRATTMRLLADPDSVADDFTKPGHVVPLRAKDGGVLR
Mvan_2694|M.vanbaalenii_PYR-1 DAKKGVGTGISAMDRATTMRLLADPDAVADDFTKPGHVVPLRAKDGGVLR
Rv1415|M.tuberculosis_H37Rv DARNGIGTGISASDRATTMRLLADPTSVADDFTRPGHVVPLRAKDGGVLR
Mb1450|M.bovis_AF2122/97 DARNGIGTGISASDRATTMRLLADPTSVADDFTRPGHVVPLRAKDGGVLR
MAV_3365|M.avium_104 DARNGVGTGISASDRATTMRLLADPTSIAEDFTRPGHVVPLRAKDGGVLR
MLBr_00559|M.leprae_Br4923 DARNGVGTGISASDRATTMRLLADPASVADDFTRPGHVVPLRAKDGGVLR
MMAR_2222|M.marinum_M DAKNGVGTGISASDRATTMRLLADPASVAEDFTRPGHVVPLRAKDGGVLR
MUL_1809|M.ulcerans_Agy99 DAKNGVGTGISASDRATTMRLLADPASVAEDFTRPGHVVPLRAKDGGVLR
MSMEG_3072|M.smegmatis_MC2_155 DAKKDVGTGISASDRATTMRALANPGSVADDFTKPGHVVPLRAKDGGVLR
TH_1011|M.thermoresistible__bu DAKHGVTTGISAADRATTMRLLADPNAVADDFTRPGHVVPLRAKEGGVLR
MAB_2796c|M.abscessus_ATCC_199 DAKNGIGTGISASDRAATMRLLADAASTADDFTKPGHVVPLRAKDGGVLR
**: .: ***** ***:*** **:. : *:***:**********:*****
Mflv_3719|M.gilvum_PYR-GCK RPGHTEAAVDLARLAGLQAAGTICEIVSQKDEGAMAQTDELRIFADEHDL
Mvan_2694|M.vanbaalenii_PYR-1 RPGHTEAAVDLARLAGLQPAGAICEIVSQKDEGAMAQTDELRIFADEHDL
Rv1415|M.tuberculosis_H37Rv RPGHTEAAVDLARMAGLQPAGAICEIVSQKDEGSMAHTDELRVFADEHGL
Mb1450|M.bovis_AF2122/97 RPGHTEAAVDLARMAGLQPAGAICEIVSQKDEGSMAHTDELRVFADEHGL
MAV_3365|M.avium_104 RPGHTEAAVDLARMAGLQPAGAICEIVSQKDEGSMAQTDELRVFADEHDL
MLBr_00559|M.leprae_Br4923 RPGHTEAAVDLARLAGLQPAGAICEIVSQKDEGSMAQTDELRVFADEHDL
MMAR_2222|M.marinum_M RPGHTEAAVDLARMAGLQPAGAICEIVSQKDEGSMAQTDELRVFADEHGL
MUL_1809|M.ulcerans_Agy99 RPGHTEAAVDLARMAGLQPAGAICEIVSRKDEGSMAQTDELRVFADEHGL
MSMEG_3072|M.smegmatis_MC2_155 RPGHTEAAVDLARLAGLQPAGAICEIVSQKDEGDMARTDELRVFADDHDL
TH_1011|M.thermoresistible__bu RPGHTEAAVDLAKLAGLQPAGAICEIVSQKDEGAMAKTDELRVFADDHGL
MAB_2796c|M.abscessus_ATCC_199 RPGHTEASVDLAKLAGLRPAAVICEIVSQKDEGAMAQTEELRVFADEHNL
*******:****::***:.*..******:**** **:*:***:***:*.*
Mflv_3719|M.gilvum_PYR-GCK ALISIADLIEWRRKHEKHIARVAEARIPTRHGEFRAVGYTSIYEDVEHVA
Mvan_2694|M.vanbaalenii_PYR-1 ALISIADLIEWRRKHEKHIARVAEARIPTRHGEFRAVGYTSIYEDVEHVA
Rv1415|M.tuberculosis_H37Rv ALITIADLIEWRRKHEKHIERVAEARIPTRHGEFRAIGYTSIYEDVEHVA
Mb1450|M.bovis_AF2122/97 ALITIADLIEWRRKHEKHIERVAEARIPTRHGEFRAIGYTSIYEDVEHVA
MAV_3365|M.avium_104 AMITIADLIEWRRKHEKHIERIAEARIPTRHGEFRAIGYTSIYEEVEHVA
MLBr_00559|M.leprae_Br4923 ALITIADLIAWRRKHEKHIERVAEARIPTRHGEFRAIGYTSIYDDVKHVA
MMAR_2222|M.marinum_M ALITIADLIEWRRKHEKHIERVAEARIPTRHGEFRAIGYTSIYEDVEHVA
MUL_1809|M.ulcerans_Agy99 ALITIADLIEWRRKHEKHIERVAEARIPTRRGEFRAIGYTSIYENVEHVA
MSMEG_3072|M.smegmatis_MC2_155 ALISIADLIEWRRKHEKHIERIAEARIPTRHGEFRAVGYKSIYEDVEHVA
TH_1011|M.thermoresistible__bu AMISIADLIEWRRKHEKHVERVAEARIPTRHGEFRAVGYTSIYDDVEHVA
MAB_2796c|M.abscessus_ATCC_199 ALVSIADLIEWRRKHEKHVQRIAEARIPTQHGEFRAVGYTSIYDDVEHVA
*:::***** ********: *:*******::*****:**.***::*:***
Mflv_3719|M.gilvum_PYR-GCK LVKGDIAGPHGDGHDVLVRVHSECLTGDVFGSRRCDCGPQLDAALAMVAR
Mvan_2694|M.vanbaalenii_PYR-1 LVKGEISGPHSDGHDVLVRVHSECLTGDVFGSRRCDCGPQLDAALAMVAR
Rv1415|M.tuberculosis_H37Rv LVRGEIAGPNADGDDVLVRVHSECLTGDVFGSRRCDCGPQLDAALAMVAR
Mb1450|M.bovis_AF2122/97 LVRGEIAGPNADGDDVLVRVHSECLTGDVFGSRRCDCGPQLDAALAMVAR
MAV_3365|M.avium_104 LVRGEIAGPNSDGDDVLVRVHSECLTGDVFGSRRCDCGPQLDAAMAMVAR
MLBr_00559|M.leprae_Br4923 LVRGEIAGSSGDGDDILVRVHSECLTGDVFGSRRCDCGTQLDAAMAMVAR
MMAR_2222|M.marinum_M LVRGEIAGPNSDGDDVLVRVHSECLTGDVFGSRRCDCGPQLDAAMAMVAR
MUL_1809|M.ulcerans_Agy99 LVRGEIAGPNSDGDDVLVRVHSECLTGDVFGSRRCDCGSQLDAAMAMVAR
MSMEG_3072|M.smegmatis_MC2_155 LVRGEISGPGSDGDDVLVRVHSECLTGDVFGSRRCDCGPQLDAAMAMVAR
TH_1011|M.thermoresistible__bu LVMGDIAGPHGDGHDVLVRVHSECLTGDVFGSRRCDCGPQLDAALSMVAR
MAB_2796c|M.abscessus_ATCC_199 LVLGDISGPDGDGNDVLVRVHSECLTGDVFGSRRCDCGPQLDAAMEMVAK
** *:*:*. .**.*:**********************.*****: ***:
Mflv_3719|M.gilvum_PYR-GCK EGRGIVLYMRGHEGRGIGLMHKLQAYQLQDAGDDTVDANLKLGLPADARD
Mvan_2694|M.vanbaalenii_PYR-1 EGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGDDTVDANLKLGLPADARD
Rv1415|M.tuberculosis_H37Rv EGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGADTVDANLKLGLPADARD
Mb1450|M.bovis_AF2122/97 EGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGADTVDANLKLGLPADARD
MAV_3365|M.avium_104 EGRGIVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGFPADARD
MLBr_00559|M.leprae_Br4923 EGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGADTVDANIKLGFPSDARD
MMAR_2222|M.marinum_M EGRGVVLYMRGHEGRGIGLLHKLQAYQLQDAGDDTVDANLKLGLPADARD
MUL_1809|M.ulcerans_Agy99 EGRGVVLYMRGHEGRGIGLLHKLQTYQLQDAGDDTVDANLKLGLPADARD
MSMEG_3072|M.smegmatis_MC2_155 EGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGLPADARD
TH_1011|M.thermoresistible__bu EGRGVVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGLPADARD
MAB_2796c|M.abscessus_ATCC_199 EGRGIVLYMRGHEGRGIGLMHKLQAYQLQDAGEDTVDANLKLGLPADARD
****:**************:****:******* ******:***:*:****
Mflv_3719|M.gilvum_PYR-GCK YGTGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAEN
Mvan_2694|M.vanbaalenii_PYR-1 YGTGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAEN
Rv1415|M.tuberculosis_H37Rv YGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAEN
Mb1450|M.bovis_AF2122/97 YGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAEN
MAV_3365|M.avium_104 YGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAEN
MLBr_00559|M.leprae_Br4923 YGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLQIIERVPLPVRANADN
MMAR_2222|M.marinum_M YGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAEN
MUL_1809|M.ulcerans_Agy99 YGIGAQILVDLGVRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANAEN
MSMEG_3072|M.smegmatis_MC2_155 YGIGAQILVDLGIRSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANKEN
TH_1011|M.thermoresistible__bu YGIGAQILVDLGVRSMRLLTNNPAKRAGLDGYGLHIIERVPLPVRATADN
MAB_2796c|M.abscessus_ATCC_199 YGLGAQILVDLGVKSMRLLTNNPAKRVGLDGYGLHIIERVPLPVRANSEN
** *********::************.*******:***********. :*
Mflv_3719|M.gilvum_PYR-GCK IRYLMTKRDRMGHDLVGLDDFDEAV----------------PGEF--GGA
Mvan_2694|M.vanbaalenii_PYR-1 IRYLMTKRDRMGHDLVGLEDFDEAV----------------PGEL--GGA
Rv1415|M.tuberculosis_H37Rv IRYLMTKRDKLGHDLAGLDDFHESVHL--------------PGEF--GGA
Mb1450|M.bovis_AF2122/97 IRYLMTKRDKLGHDLAGLDDFHESVHL--------------PGEF--GGA
MAV_3365|M.avium_104 IRYLMTKRDKMGHDLAGLDDFHESVHL--------------PGEF--GGA
MLBr_00559|M.leprae_Br4923 IRYLMTKRDKMGHDLGGLDDFHESNYF--------------PGEF--GGA
MMAR_2222|M.marinum_M IRYLMTKRDRMGHDLTGLDDFHESVHL--------------PGEF--GGA
MUL_1809|M.ulcerans_Agy99 IRYLMTKRDRMGHDLTGLDDFHESVHL--------------PGEF--GGA
MSMEG_3072|M.smegmatis_MC2_155 IRYLMTKRDRMGHDLVGLDDYDEAVSMDDYDEAVYLLGDRRPTES--GGA
TH_1011|M.thermoresistible__bu IRYLMTKRDRMGHDLVGLEDFDESVRMDDYDEGVYLLGDRRPTDLDLGGA
MAB_2796c|M.abscessus_ATCC_199 IRYLRTKRDRMGHDLADLDDHPEAD-----------------------GA
**** ****::**** .*:*. *: **
Mflv_3719|M.gilvum_PYR-GCK V
Mvan_2694|M.vanbaalenii_PYR-1 V
Rv1415|M.tuberculosis_H37Rv L
Mb1450|M.bovis_AF2122/97 L
MAV_3365|M.avium_104 L
MLBr_00559|M.leprae_Br4923 L
MMAR_2222|M.marinum_M L
MUL_1809|M.ulcerans_Agy99 L
MSMEG_3072|M.smegmatis_MC2_155 Q
TH_1011|M.thermoresistible__bu L
MAB_2796c|M.abscessus_ATCC_199 -