For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MYADRDLPGAGGLAVRVIPCLDVDDGRVVKGVNFENLRDAGDPVELAAVYDAEGADELTFLDVTASSSGR ATMLEVVRRTAEQVFIPLTVGGGVRTVADVDSLLRAGADKVAVNTAAIACPDLLADMARQFGSQCIVLSV DARTVPVGSAPTPSGWEVTTHGGRRGTGMDAVQWAARGADLGVGEILLNSMDADGTKAGFDLALLRAVRA AVTVPVIASGGAGAVEHFAPAVAAGADAVLAASVFHFRELTIGQVKAALAAEGITVR
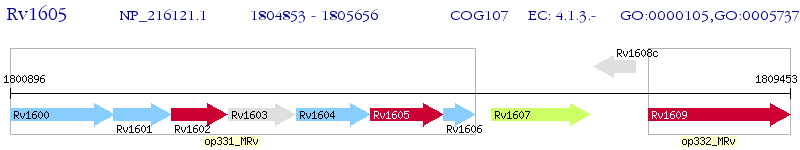
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1605 | hisF | - | 100% (267) | imidazole glycerol phosphate synthase subunit HisF |
| M. tuberculosis H37Rv | Rv1603 | hisA | 4e-15 | 25.40% (248) | phosphoribosyl isomerase A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1631 | hisF | 1e-147 | 99.63% (267) | imidazole glycerol phosphate synthase subunit HisF |
| M. gilvum PYR-GCK | Mflv_3607 | - | 1e-125 | 86.82% (258) | imidazole glycerol phosphate synthase subunit HisF |
| M. leprae Br4923 | MLBr_01263 | hisF | 1e-125 | 89.84% (256) | imidazole glycerol phosphate synthase subunit HisF |
| M. abscessus ATCC 19977 | MAB_2664c | - | 1e-119 | 84.31% (255) | imidazole glycerol phosphate synthase, cyclase subunit HisF |
| M. marinum M | MMAR_2401 | hisF | 1e-132 | 94.14% (256) | cyclase HisF |
| M. avium 104 | MAV_3181 | hisF | 1e-130 | 89.62% (260) | imidazole glycerol phosphate synthase subunit HisF |
| M. smegmatis MC2 155 | MSMEG_3211 | hisF | 1e-124 | 87.06% (255) | imidazole glycerol phosphate synthase subunit HisF |
| M. thermoresistible (build 8) | TH_1157 | hisF | 1e-121 | 85.88% (255) | Probable cyclase hisF |
| M. ulcerans Agy99 | MUL_1578 | hisF | 1e-132 | 93.36% (256) | imidazole glycerol phosphate synthase subunit HisF |
| M. vanbaalenii PYR-1 | Mvan_2810 | - | 1e-127 | 88.76% (258) | imidazole glycerol phosphate synthase subunit HisF |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3607|M.gilvum_PYR-GCK ---MKPA------SDVATRVIPCLDVDDGRVVKGVNFANLRDAGDPVELA
Mvan_2810|M.vanbaalenii_PYR-1 ---MKPA------GDVATRVIPCLDVDGGRVVKGVNFANLRDAGDPVELA
MAB_2664c|M.abscessus_ATCC_199 ---MS----------VATRVIACLDVDDGRVVKGVNFENLRDAGDPVELA
MSMEG_3211|M.smegmatis_MC2_155 ---MSAN------SDVATRVIPCLDVDNGRVVKGVNFENLRDAGDPVELA
TH_1157|M.thermoresistible__bu MTDNTAD------RKLAVRVIPCLDVDAGRVVKGVRFENLRDAGDPVELA
Rv1605|M.tuberculosis_H37Rv ---MYADRDLPGAGGLAVRVIPCLDVDDGRVVKGVNFENLRDAGDPVELA
Mb1631|M.bovis_AF2122/97 ---MYADRDLPGAGGLAVRVIPCLDVDDGRVVKGVNFENLRDAGDPVELA
MMAR_2401|M.marinum_M ---MYTD------TGLAVRVIPCLDVDAGRVVKGVNFENLRDAGDPVELA
MUL_1578|M.ulcerans_Agy99 ---MYTD------TGLAVRVIPCLDIDAGRVVKGVNFENLRDAGDPVELA
MLBr_01263|M.leprae_Br4923 ---MYSG------NGLAVRVIPCLDVYCGRVVKGVNFKNLRDAGDLVELA
MAV_3181|M.avium_104 ---MSPN-----SSGLAVRVIPCLDVDDGRVVKGVNFENLRDAGDPVELA
:*.***.***: *******.* ******* ****
Mflv_3607|M.gilvum_PYR-GCK AAYDAEGADELTFLDVTASSSGRSTMLEVVRRTAEQVFIPLTVGGGVRSI
Mvan_2810|M.vanbaalenii_PYR-1 AAYDAEGADELTFLDVTASSSGRATMLDVVRRTAEQVFIPLTVGGGVRSV
MAB_2664c|M.abscessus_ATCC_199 AAYDAEGVDELTFLDVTASSSGRATMLDVVRRTAEQVFIPLTVGGGVRSV
MSMEG_3211|M.smegmatis_MC2_155 AAYDAEGADELTFLDVTASSSGRSTMLEVVRRTAEQVFIPLTVGGGVRSV
TH_1157|M.thermoresistible__bu AVYDAEGADELCFLDVTASSSGRSTMLDVVRRTAEQVFIPLTVGGGVRSV
Rv1605|M.tuberculosis_H37Rv AVYDAEGADELTFLDVTASSSGRATMLEVVRRTAEQVFIPLTVGGGVRTV
Mb1631|M.bovis_AF2122/97 AVYDAEGADELTFLDVTASSSGRATMLEVVRRTAEQVFIPLTVGGGVRTV
MMAR_2401|M.marinum_M AAYDAEGADELTFLDVTASSSGRATMLEVVRHTAEQVFIPLTVGGGVRTV
MUL_1578|M.ulcerans_Agy99 AAYDAEGADELTFLDVTASSSGRATMLEVVRHTAEQVFIPLTVGGGVRTV
MLBr_01263|M.leprae_Br4923 AAYDAEGADELAFLDVTASSSGRATMLEVVRCTAEQVFIPLMVGGGVRTV
MAV_3181|M.avium_104 AVYDAEGADELTFLDVTASSSGRATMLDVVRRTAEQVFIPLTVGGGVRTV
*.*****.*** ***********:***:*** ********* ******::
Mflv_3607|M.gilvum_PYR-GCK ADVDVLLRAGADKVSVNTAAIARPELLAELSRQFGSQCIVLSVDARTVPE
Mvan_2810|M.vanbaalenii_PYR-1 ADVDALLRAGADKVSVNTAAIARPELLSELSRQFGSQCIVLSVDARTVPE
MAB_2664c|M.abscessus_ATCC_199 DDVNVLLRAGADKVGVNTAAIARPELLAELAQRFGSQCIVLSVDARRVRD
MSMEG_3211|M.smegmatis_MC2_155 DDVDVLLRAGADKVSVNTAAIARPELLAEMARQFGSQCIVLSVDARTVPK
TH_1157|M.thermoresistible__bu ADVDTLLRAGADKVSVNTAAIERPELLSELARQFGSQCIVLSVDARTVPE
Rv1605|M.tuberculosis_H37Rv ADVDSLLRAGADKVAVNTAAIACPDLLADMARQFGSQCIVLSVDARTVPV
Mb1631|M.bovis_AF2122/97 ADVDSLLRAGADKVAVNTAAIACLDLLADMARQFGSQCIVLSVDARTVPV
MMAR_2401|M.marinum_M ADVDLLLRAGADKVSVNTAAIARPDLLADMATQFGSQCIVLSVDARKVPV
MUL_1578|M.ulcerans_Agy99 ADVDLLLRAGADKVSVNTAAIARPDLLADMATQFGSQCIVLSVDARKVPV
MLBr_01263|M.leprae_Br4923 ADVDVLLRAGADKVAVNTAAIARPELLADMAGQFGSQCIVLSVDARTVPT
MAV_3181|M.avium_104 ADVDVLLRAGADKVSVNTAAIARPELLEEMARQFGSQCIVLSVDARTVPP
**: *********.****** :** ::: :************* *
Mflv_3607|M.gilvum_PYR-GCK GSTPTTSGWEVTTHGGRRGTGIDAVEWASQGAELGVGEILLNSMDADGTK
Mvan_2810|M.vanbaalenii_PYR-1 GSQPTPSGWEVTTHGGRRGTGIDAVEWAVQGAELGVGEILLNSMDADGTK
MAB_2664c|M.abscessus_ATCC_199 GDVPTSSGWEVTTHGGRRGTGIDAIEWTSRGAELGVGEILLNSMDADGTK
MSMEG_3211|M.smegmatis_MC2_155 GEQPTPSGWEVTTHGGRRGTGIDAIEWATRGAELGVGEILLNSMDRDGTK
TH_1157|M.thermoresistible__bu GSPPTSSGWEVTTHGGRRGTGIDAVEWAVRGAELGVGEILLNSMDRDGTK
Rv1605|M.tuberculosis_H37Rv GSAPTPSGWEVTTHGGRRGTGMDAVQWAARGADLGVGEILLNSMDADGTK
Mb1631|M.bovis_AF2122/97 GSAPTPSGWEVTTHGGRRGTGMDAVQWAARGADLGVGEILLNSMDADGTK
MMAR_2401|M.marinum_M GPAPTPSGWEVTTHGGRRGTGIDAVEWAARGADLGVGEILLNSMDADGTK
MUL_1578|M.ulcerans_Agy99 GSSPTPSGWEVTTHGGRRGTGIDAVEWAARGADLGVGEILLNSMDADGTK
MLBr_01263|M.leprae_Br4923 GSARTPSGWEATTHGGYRGTGIDAVEWAARGADLGVGEILLNSMDADGTK
MAV_3181|M.avium_104 GAVPTPSGWEVTTHGGRRGTGIDAVEWASRGADLGVGEILLNSMDADGTK
* *.****.***** ****:**::*: :**:************ ****
Mflv_3607|M.gilvum_PYR-GCK AGFDLKMLRAVRGAVTVPVIASGGAGAVEHFAPAVFAGADAVLAASVFHF
Mvan_2810|M.vanbaalenii_PYR-1 AGFDLEMLRAVRGAVTVPVIASGGAGAVEHFAPAVAAGADAVLAASVFHF
MAB_2664c|M.abscessus_ATCC_199 AGFDLEMIAAARAAVDVPVIASGGAGAIGHFAPAVQAGADAVLAASVFHF
MSMEG_3211|M.smegmatis_MC2_155 AGFDLKMLSAVRAAVSVPVIASGGAGAVEHFAPAVQAGADAVLAASVFHF
TH_1157|M.thermoresistible__bu DGFDLPMLRAVRGAVSIPIIASGGAGALEHFPAAVHAGADAVLAASVFHF
Rv1605|M.tuberculosis_H37Rv AGFDLALLRAVRAAVTVPVIASGGAGAVEHFAPAVAAGADAVLAASVFHF
Mb1631|M.bovis_AF2122/97 AGFDLALLRAVRAAVTVPVIASGGAGAVEHFAPAVAAGADAVLAASVFHF
MMAR_2401|M.marinum_M AGFDLAMLRAVRAAVTVPVIASGGAGNVDHFAPAVAAGADAVLAASVFHF
MUL_1578|M.ulcerans_Agy99 AGFDLAMLRAVRAAVTVPVIASGGAGNVDHFAPAVAAGADAVLAASVFHF
MLBr_01263|M.leprae_Br4923 AGFDLAMLRAVRAAVTVPVIASGGAGAIEHFVPAVTAGADAVLAASVFHF
MAV_3181|M.avium_104 AGFDLEMLQAVRSAVTVPVIASGGAGAAEHFAPAIEAGADAVLAASVFHF
**** :: *.*.** :*:******* ** .*: **************
Mflv_3607|M.gilvum_PYR-GCK GDLTIGEVKAAMKAEGIVVR---
Mvan_2810|M.vanbaalenii_PYR-1 GELTIGQVKAAMKAEGITVR---
MAB_2664c|M.abscessus_ATCC_199 RELTIGEVKAAMAAEGITVR---
MSMEG_3211|M.smegmatis_MC2_155 RELTVAEVKASMKAAGITVR---
TH_1157|M.thermoresistible__bu RELTIKQVKEAMAAEGITVR---
Rv1605|M.tuberculosis_H37Rv RELTIGQVKAALAAEGITVR---
Mb1631|M.bovis_AF2122/97 RELTIGQVKAALAAEGITVR---
MMAR_2401|M.marinum_M RELTIGQVKAAMAAEGITVR---
MUL_1578|M.ulcerans_Agy99 RELTIGQVKAAMAAEGNTVR---
MLBr_01263|M.leprae_Br4923 RELTIGQVKDAMAAAGIAVR---
MAV_3181|M.avium_104 RELTIGQVKAAMAEAGIPVRMVR
:**: :** :: * **