For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAQHNIVVIGGGGAGLRAAIAIAETNPHLDVAIVSKVYPMRSHTVSAEGGAAAVTGDDDSLDEHAHDTV SGGDWLCDQDAVEAFVAEAPKELVQLEHWGCPWSRKPDGRVAVRPFGGMKKLRTWFAADKTGFHLLHTLF QRLLTYSDVMRYDEWFATTLLVDDGRVCGLVAIELATGRIETILADAVILCTGGCGRVFPFTTNANIKTG DGMALAFRAGAPLKDMEFVQYHPTGLPFTGILITEAARAEGGWLLNKDGYRYLQDYDLGKPTPEPRLRSM ELGPRDRLSQAFVHEHNKGRTVDTPYGPVVYLDLRHLGADLIDAKLPFVRELCRDYQHIDPVVELVPVRP VVHYMMGGVHTDINGATTLPGLYAAGETACVSINGANRLGSNSLPELLVFGARAGRAAADYAARHQKSDR GPSSAVRAQARTEALRLERELSRHGQGGERIADIRADMQATLESAAGIYRDGPTLTKAVEEIRVLQERFA TAGIDDHSRTFNTELTALLELSGMLDVALAIVESGLRREESRGAHQRTDFPNRDDEHFLAHTLVHRESDG TLRVGYLPVTITRWPPGERVYGR
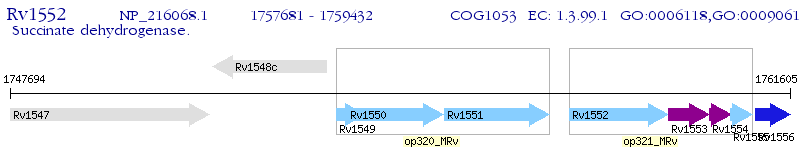
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1552 | frdA | - | 100% (583) | fumarate reductase flavoprotein subunit |
| M. tuberculosis H37Rv | Rv3318 | sdhA | e-128 | 42.41% (606) | succinate dehydrogenase flavoprotein subunit |
| M. tuberculosis H37Rv | Rv0248c | sdhA | 2e-81 | 36.05% (577) | succinate dehydrogenase flavoprotein subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1578 | frdA | 0.0 | 100.00% (583) | fumarate reductase flavoprotein subunit |
| M. gilvum PYR-GCK | Mflv_4846 | sdhA | 1e-129 | 43.17% (600) | succinate dehydrogenase flavoprotein subunit |
| M. leprae Br4923 | MLBr_00697 | sdhA | 1e-127 | 42.33% (600) | succinate dehydrogenase flavoprotein subunit |
| M. abscessus ATCC 19977 | MAB_3675 | sdhA | 1e-129 | 43.00% (600) | succinate dehydrogenase flavoprotein subunit |
| M. marinum M | MMAR_1201 | sdhA | 1e-128 | 43.00% (600) | succinate dehydrogenase (flavoprotein subunit) SdhA |
| M. avium 104 | MAV_4299 | sdhA | 1e-128 | 43.00% (600) | succinate dehydrogenase flavoprotein subunit |
| M. smegmatis MC2 155 | MSMEG_1670 | sdhA | 1e-128 | 42.50% (600) | succinate dehydrogenase flavoprotein subunit |
| M. thermoresistible (build 8) | TH_2182 | sdhA | 1e-130 | 43.50% (600) | PROBABLE SUCCINATE DEHYDROGENASE (FLAVOPROTEIN SUBUNIT) SDHA |
| M. ulcerans Agy99 | MUL_1369 | sdhA | 1e-127 | 42.83% (600) | succinate dehydrogenase flavoprotein subunit |
| M. vanbaalenii PYR-1 | Mvan_1587 | sdhA | 1e-128 | 43.00% (600) | succinate dehydrogenase flavoprotein subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4846|M.gilvum_PYR-GCK ----MIVEHRYDVVIVGAGGAGMRAAV---EAGPRVRTAVLTKLYPTRSH
Mvan_1587|M.vanbaalenii_PYR-1 ----MIVEHRYDVVIVGAGGAGMRAAV---EAGPRARTAVLTKLYPTRSH
MSMEG_1670|M.smegmatis_MC2_155 ----MIQEHRYDVVIVGAGGAGMRAAV---EAGPRARTAVLTKLYPTRSH
MAB_3675|M.abscessus_ATCC_1997 MGSPIIQEHRYDVVIIGAGGAGMRAAV---EAGPRARTAVLTKLYPTRSH
TH_2182|M.thermoresistible__bu ----MIQEHRYDVVIVGAGGAGMRAAV---EAGPRVRTAVLTKVYPTRSH
MLBr_00697|M.leprae_Br4923 ----MIQQHRYDVVIVGAGGAGMRAAV---EAGPRVRTAVLTKLYPTRSH
MAV_4299|M.avium_104 ----MIQQHRYDVVIVGAGGAGMRAAV---EAGPRVRTAVLTKLYPTRSH
MMAR_1201|M.marinum_M ----MIQQHRYDVVIVGAGGAGMRAAV---EAGPRVRTAVLTKLYPTRSH
MUL_1369|M.ulcerans_Agy99 ----MIQQHRYDVVIVGAGGAGMRAAV---EAGPRVRTAVLAKLYPTRSH
Rv1552|M.tuberculosis_H37Rv ----MTAQH--NIVVIGGGGAGLRAAIAIAETNPHLDVAIVSKVYPMRSH
Mb1578|M.bovis_AF2122/97 ----MTAQH--NIVVIGGGGAGLRAAIAIAETNPHLDVAIVSKVYPMRSH
: :* ::*::*.****:***: *:.*: .*:::*:** ***
Mflv_4846|M.gilvum_PYR-GCK TGAAQGGMCAALANVEEDNWEWHTFDTVKGGDYLADQDAVEIMAKEAIDA
Mvan_1587|M.vanbaalenii_PYR-1 TGAAQGGMCAALANVEEDNWEWHTFDTVKGGDYLADQDAVEIMAKEAIDA
MSMEG_1670|M.smegmatis_MC2_155 TGAAQGGMCAALANVEEDNWEWHTFDTVKGGDYLADQDAVEIMCKEAIDA
MAB_3675|M.abscessus_ATCC_1997 TGAAQGGMCAALANVEEDNWEWHTFDTVKGGDYLADQDAVEIMCKEAIDA
TH_2182|M.thermoresistible__bu TGAAQGGMCAALANVEEDNWEWHAFDTVKGGDYLADQDAVEIMCREAIDA
MLBr_00697|M.leprae_Br4923 TGAAQGGMCAALANVEDDNWEWHAFDTVKGGDYLADQDAVEIMCKEAIDA
MAV_4299|M.avium_104 TGAAQGGMCAALANVEEDNWEWHTFDTVKGGDYLADQDAVEIMCKEAIDA
MMAR_1201|M.marinum_M TGAAQGGMCAALANVEDDNWEWHTFDTVKGGDYLADQDAVEIMCKEAIDA
MUL_1369|M.ulcerans_Agy99 TGAAQGGMCAALANVEDDNWEWHTFDTVKGGDYLADQDAVEIMCKEAIDA
Rv1552|M.tuberculosis_H37Rv TVSAEGGAAAVTG--DDDSLDEHAHDTVSGGDWLCDQDAVEAFVAEAPKE
Mb1578|M.bovis_AF2122/97 TVSAEGGAAAVTG--DDDSLDEHAHDTVSGGDWLCDQDAVEAFVAEAPKE
* :*:** .*. . ::*. : *:.***.***:*.****** : ** .
Mflv_4846|M.gilvum_PYR-GCK VLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
Mvan_1587|M.vanbaalenii_PYR-1 VLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
MSMEG_1670|M.smegmatis_MC2_155 VLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
MAB_3675|M.abscessus_ATCC_1997 VLDLEKMGMPFNRTPQGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
TH_2182|M.thermoresistible__bu VLDLEKMGMPFNRTPEGKIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
MLBr_00697|M.leprae_Br4923 VLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
MAV_4299|M.avium_104 VLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
MMAR_1201|M.marinum_M VLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
MUL_1369|M.ulcerans_Agy99 VLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHMI
Rv1552|M.tuberculosis_H37Rv LVQLEHWGCPWSRKPDGRVAVRPFGGMKK-------LRTWFAADKTGFHL
Mb1578|M.bovis_AF2122/97 LVQLEHWGCPWSRKPDGRVAVRPFGGMKK-------LRTWFAADKTGFHL
:::**: * *:.*.*:*:: * *** .: *: :***:**. :
Mflv_4846|M.gilvum_PYR-GCK LQTLYQNCVKH-DVEFFNEFYALDITMTETPSGPVATGVVAYELATGDIH
Mvan_1587|M.vanbaalenii_PYR-1 LQTLYQNCVKH-DVEFFNEFYALDITLTETASGPVATGVVAYELATGDIH
MSMEG_1670|M.smegmatis_MC2_155 LQTLYQNCVKH-DVEFFNEFYALDIALTETPAGPVATGVIAYELATGDIH
MAB_3675|M.abscessus_ATCC_1997 LQTLYQNCVKH-DVEFFNEFYALDIALTETPNGPVATGVVAYELATGDIH
TH_2182|M.thermoresistible__bu LQTLYQNCVKH-DVQFFNEFYALDLAMTETPSGRVATGVIAYELATGDIH
MLBr_00697|M.leprae_Br4923 LQTLYQNCVKH-DVEFFNEFYALDLVLTQTPSGPVTTGVVAYELATGDIH
MAV_4299|M.avium_104 LQTLYQNCVRH-DVQFFNEFYALDLVLTQTPSGPVASGVVAYELATGDIH
MMAR_1201|M.marinum_M LQTLYQNCVKH-DVEFFNEFYALDLALTETPSGPVATGVIAYELATGDIH
MUL_1369|M.ulcerans_Agy99 LQTLYQNCVKH-DVEFFNEFYALDLALTETPSGPVATGVIAYELATGDIH
Rv1552|M.tuberculosis_H37Rv LHTLFQRLLTYSDVMRYDEWFATTLLVDDGR----VCGLVAIELATGRIE
Mb1578|M.bovis_AF2122/97 LHTLFQRLLTYSDVMRYDEWFATTLLVDDGR----VCGLVAIELATGRIE
*:**:*. : : ** ::*::* : : : . *::* ***** *.
Mflv_4846|M.gilvum_PYR-GCK VFHAKAIVFATGGSGRMYKTTSNAHTLTGDGLGIIFRKGLPLEDMEFHQF
Mvan_1587|M.vanbaalenii_PYR-1 IFHAKAIVFATGGSGRMYKTTSNAHTLTGDGLGIIFRKGLPLEDMEFHQF
MSMEG_1670|M.smegmatis_MC2_155 VFHAKAIVFATGGSGRMYKTTSNAHTLTGDGLGIVFRKGLPLEDMEFHQF
MAB_3675|M.abscessus_ATCC_1997 VFHAKAIVFATGGSGRMYKTTSNAHTLTGDGLGIVFRKGLPLEDMEFHQF
TH_2182|M.thermoresistible__bu VFHAKAIVFATGGSGRMYKTTSNAHTLTGDGLGIVFRKGLPLEDMEFHQF
MLBr_00697|M.leprae_Br4923 VFHTKAVVIATGGSGRMYKTTSNAHTLTGDGIGIVFRKGLPLEDMEFHQF
MAV_4299|M.avium_104 VFHAKAVVLATGGSGRMYKTTSNAHTLTGDGIGIVFRKGLPLEDMEFHQF
MMAR_1201|M.marinum_M IFHAKAIVLATGGSGRMYKTTSNAHTLTGDGIGIVFRKGLPLEDMEFHQF
MUL_1369|M.ulcerans_Agy99 IFHAKAIVLATGGSGRMYKTTSNAHTLTGDGIGIVFRKGLPLEDMEFHQF
Rv1552|M.tuberculosis_H37Rv TILADAVILCTGGCGRVFPFTTNANIKTGDGMALAFRAGAPLKDMEFVQY
Mb1578|M.bovis_AF2122/97 TILADAVILCTGGCGRVFPFTTNANIKTGDGMALAFRAGAPLKDMEFVQY
: :.*:::.***.**:: *:**: ****:.: ** * **:**** *:
Mflv_4846|M.gilvum_PYR-GCK HPTGLAGLGILISEAVRGEGGRLLNGDGERFMERYAP---------TIVD
Mvan_1587|M.vanbaalenii_PYR-1 HPTGLAGLGILISEAVRGEGGRLLNGEGERFMERYAP---------TIVD
MSMEG_1670|M.smegmatis_MC2_155 HPTGLAGLGILISEAVRGEGGRLLNGEGERFMERYAP---------TIVD
MAB_3675|M.abscessus_ATCC_1997 HPTGLAGLGILISEAVRGEGGRLLNADGERFMERYAP---------TIVD
TH_2182|M.thermoresistible__bu HPTGLAGLGILISEAVRGEGGRLLNGEGERFMERYAP---------TIVD
MLBr_00697|M.leprae_Br4923 HPTGLAGLGILISEAVRGEGGRLLNGENERFMEHYAP---------TIVD
MAV_4299|M.avium_104 HPTGLAGLGILISEAVRGEGGRLLNGEGERFMERYAP---------TIVD
MMAR_1201|M.marinum_M HPTGLAGLGILISEAVRGEGGRLLNGEGERFMERYAP---------TIVD
MUL_1369|M.ulcerans_Agy99 HPTGLAGLGILISEAVRGEGGRLLNGEGERFMERYAP---------TIVD
Rv1552|M.tuberculosis_H37Rv HPTGLPFTGILITEAARAEGGWLLNKDGYRYLQDYDLGKPTPEPRLRSME
Mb1578|M.bovis_AF2122/97 HPTGLPFTGILITEAARAEGGWLLNKDGYRYLQDYDLGKPTPEPRLRSME
*****. ****:**.*.*** *** :. *::: * ::
Mflv_4846|M.gilvum_PYR-GCK LAPRDIVARSMVLEVLEGRGAG-PNKDYVYIDVRHLGEDVLEAKLPDITE
Mvan_1587|M.vanbaalenii_PYR-1 LAPRDIVARSMVLEVLEGRGAG-PNKDYVYIDVRHLGEDVLEAKLPDITE
MSMEG_1670|M.smegmatis_MC2_155 LAPRDIVARSMVLEVLEGRGAG-PNKDYVYIDVRHLGEDVLEAKLPDITE
MAB_3675|M.abscessus_ATCC_1997 LAPRDIVARSMVLEVLEGRGAG-PNKDYVYIDVRHLGADVLEAKLPDITE
TH_2182|M.thermoresistible__bu LAPRDIVARSMVLEVLEGRGAG-PNKDYVYIDVRHLGEDVLEAKLPDITE
MLBr_00697|M.leprae_Br4923 LAPRDIVARSMVLEVLEGRGAG-PHKDYVYIDVRHLGEEVLESKLPDITE
MAV_4299|M.avium_104 LAPRDIVARSMVLEVLEGRGAG-PHKDYVYIDVRHLGEDVLESKLPDITE
MMAR_1201|M.marinum_M LAPRDIVARSMVLEVLEGRGAG-PHKDYVYIDVRHLGEDVLEAKLPDITE
MUL_1369|M.ulcerans_Agy99 LAPRDIVARSMVLEVLEGRGAG-PHKDYVYIDVRHLGEDVLEAKLPDITE
Rv1552|M.tuberculosis_H37Rv LGPRDRLSQAFVHEHNKGRTVDTPYGPVVYLDLRHLGADLIDAKLPFVRE
Mb1578|M.bovis_AF2122/97 LGPRDRLSQAFVHEHNKGRTVDTPYGPVVYLDLRHLGADLIDAKLPFVRE
*.*** :::::* * :** .. * **:*:**** :::::*** : *
Mflv_4846|M.gilvum_PYR-GCK FARTYLGVDPVKELVPVYPTCHYVMGGIPTTVNGQVLSDNTTVVPGLYAA
Mvan_1587|M.vanbaalenii_PYR-1 FARTYLGVDPVKELVPVYPTCHYVMGGIPTTVNGQVLADNTTIIPGLYAA
MSMEG_1670|M.smegmatis_MC2_155 FARTYLGVDPVKELVPVYPTCHYVMGGIPTTVNGQVLRDNTNVIPGLYAA
MAB_3675|M.abscessus_ATCC_1997 FARTYLGVDPVTELVPVYPTCHYVMGGIPTNIHGQVLRDNDNVVPGLYAA
TH_2182|M.thermoresistible__bu FARTYLGVDPVTELVPVYPTCHYLMGGIPTTVHGQVLADNTTIVPGLYAA
MLBr_00697|M.leprae_Br4923 FSRTYLGVDPVHELVPVYPTCHYVMGGIPTTVTGQVLRDNTSTVPGLYAA
MAV_4299|M.avium_104 FARTYLGVDPVHELVPVYPTCHYVMGGIPTTVTGQVLRDNTSTVPGLYAA
MMAR_1201|M.marinum_M FARTYLGVDPVHELVPVYPTCHYVMGGIPTTVTGQVLRDNTHVVPGLFAA
MUL_1369|M.ulcerans_Agy99 FARTYLGVDPVHELVPVYPTCHYVMGGIPTTVTGQVLRDNTHVVPGLFAA
Rv1552|M.tuberculosis_H37Rv LCRDYQHIDPVVELVPVRPVVHYMMGGVHTDING------ATTLPGLYAA
Mb1578|M.bovis_AF2122/97 LCRDYQHIDPVVELVPVRPVVHYMMGGVHTDING------ATTLPGLYAA
:.* * :*** ***** *. **:***: * : * :***:**
Mflv_4846|M.gilvum_PYR-GCK GECACVSVHGANRLGTNSLLDINVFGRRAGIAAANYALGHDFVELPESPA
Mvan_1587|M.vanbaalenii_PYR-1 GECACVSVHGANRLGTNSLLDINVFGRRAGIAAANYALGHDFVDLPENPA
MSMEG_1670|M.smegmatis_MC2_155 GECACVSVHGANRLGTNSLLDINVFGRRAGIAAAEYAQNHNFVDMPENPA
MAB_3675|M.abscessus_ATCC_1997 GECACVSVHGANRLGTNSLLDINVFGRRAGIAAAEYAEAHDFIDLPENPA
TH_2182|M.thermoresistible__bu GECACVSVHGANRLGTNSLLDINVFGRRAGLAAADYAQNHDFVPLPDNPA
MLBr_00697|M.leprae_Br4923 GECACVSVHGANRLGTNSLLDINVFGRRSGIAAASYASANDFVDMPPNPA
MAV_4299|M.avium_104 GECACVSVHGANRLGTNSLLDINVFGRRAGIAAANYARGHDFVDMPPDPA
MMAR_1201|M.marinum_M GECACVSVHGANRLGTNSLLDINVFGRRAGIAAARYARGHDFVDMPAEPA
MUL_1369|M.ulcerans_Agy99 GECACVSVHGANRLGTNSLLDINVFGRRAGIAAARYARGHDFVDMPAEPA
Rv1552|M.tuberculosis_H37Rv GETACVSINGANRLGSNSLPELLVFGARAGRAAADYAARHQKSDRGPSSA
Mb1578|M.bovis_AF2122/97 GETACVSINGANRLGSNSLPELLVFGARAGRAAADYAARHQKSDRGPSSA
** ****::******:*** :: *** *:* *** ** :: ..*
Mflv_4846|M.gilvum_PYR-GCK EMVVG-----WVGDILSEHGN--ERVADIRGALQQSMDNNAAVFRTEETL
Mvan_1587|M.vanbaalenii_PYR-1 EMVVG-----WVGDILSEHGN--ERVADIRGALQQSMDNNAAVFRTEETL
MSMEG_1670|M.smegmatis_MC2_155 EMVVG-----WVGDILSEHGN--ERVADIRGALQQSMDNNAAVFRTEETL
MAB_3675|M.abscessus_ATCC_1997 TMVTE-----WVADVLSEHGD--ERVADIRTELQQSMDNNAAVFRTEDTL
TH_2182|M.thermoresistible__bu GMVTE-----WIAHILSEHGH--ERVADIRGALQQTMDNNAAVFRTEETL
MLBr_00697|M.leprae_Br4923 EMVIS-----WVANILSEHGN--ERVADIRGALQQTMDNNAAVFRTEETL
MAV_4299|M.avium_104 AMVVR-----WVADILSEHGN--ERVADIRGTLQQSMDNNAAVFRTEETL
MMAR_1201|M.marinum_M SMVVS-----WVADILSEHGT--ERVADIRGALQQTMDNNAAVFRTEETL
MUL_1369|M.ulcerans_Agy99 SMVVS-----WVADILSEHGN--EPVADIRGALQQTMDNNAAVFRTEETL
Rv1552|M.tuberculosis_H37Rv VRAQARTEALRLERELSRHGQGGERIADIRADMQATLESAAGIYRDGPTL
Mb1578|M.bovis_AF2122/97 VRAQARTEALRLERELSRHGQGGERIADIRADMQATLESAAGIYRDGPTL
. : **.** * :**** :* :::. *.::* **
Mflv_4846|M.gilvum_PYR-GCK KQALTDIHALKERYSRITVQDKGKRYNSDLLEAIELGFLLELAEVTVVGA
Mvan_1587|M.vanbaalenii_PYR-1 KQALTDIHALKERYSRITVQDKGKRYNSDLLEAIELGFLLELAEVTVVGA
MSMEG_1670|M.smegmatis_MC2_155 KQALTDIHALKERYSRITVHDKGKRYNSDLLEAIELGFLLELAEVTVVGA
MAB_3675|M.abscessus_ATCC_1997 KQALNDIHALKERYSRITVHDKGKRYNSDLLEAIELGFLLELAEVTVVGA
TH_2182|M.thermoresistible__bu KTALSDIHRLKERYSRITVQDKGKRYNSDLLEAIELGFLLELAEVTVVGA
MLBr_00697|M.leprae_Br4923 KQALTDIHSLKERYSRITVHDKGKSFNSDLLEAIELGFLLELAEVTVVGA
MAV_4299|M.avium_104 KQALTDIHACKERYSRIRIHDKGKRFNSDLLEAIELGFLLELAEVTVVGA
MMAR_1201|M.marinum_M KQALTDIHALKERYSRITVHDKGKRFNSDLLEAIELGFLLELAEVTVVGA
MUL_1369|M.ulcerans_Agy99 KQALTDIHALKERYSRITVHDKGKRFNSDLLEAIELGFLLELAEVTVVGA
Rv1552|M.tuberculosis_H37Rv TKAVEEIRVLQERFATAGIDDHSRTFNTELTALLELSGMLDVALAIVESG
Mb1578|M.bovis_AF2122/97 TKAVEEIRVLQERFATAGIDDHSRTFNTELTALLELSGMLDVALAIVESG
. *: :*: :**:: :.*:.: :*::* :**. :*::* . * ..
Mflv_4846|M.gilvum_PYR-GCK LNRKESRGGHAREDYPNRDDTNYMRHTMAYKEGSDLLSDIRLDYKPVVMT
Mvan_1587|M.vanbaalenii_PYR-1 LNRKESRGGHAREDYPNRDDTNYMRHTMAYKEGTDLLSDIRLDYKPVVQT
MSMEG_1670|M.smegmatis_MC2_155 LNRKESRGGHAREDYPNRDDTNYMRHTMAYKQGTDLLSDIRLDYKPVVQT
MAB_3675|M.abscessus_ATCC_1997 LNRKESRGGHAREDYPNRDDTNYMRHTMAYKQGAELLSDIRLDYKPVVQT
TH_2182|M.thermoresistible__bu LNRKETRGGHAREDYPNRDDTNYMRHTMAYKQGTELLSDIRLDYKPVVQT
MLBr_00697|M.leprae_Br4923 LNRKESRGGHAREDYPHRDDTNYMRHTMAYKQGTDLLSDIALDFKPVVQT
MAV_4299|M.avium_104 LNRKESRGGHAREDYPNRDDVNYMRHTMAYKQGTDLLSDIALDFKPVVQT
MMAR_1201|M.marinum_M LNRKESRGGHAREDYPNRDDVNYMRHTMAYKEGTDLLSDIRLDFKPVVQT
MUL_1369|M.ulcerans_Agy99 LNRKESRGGHAREDYPNRDDVNYMRHTMAYKEGTDLLSDIRLDFKPVVQT
Rv1552|M.tuberculosis_H37Rv LRREESRGAHQRTDFPNRDDEHFLAHTLVHRESD---GTLRVGYLPVTIT
Mb1578|M.bovis_AF2122/97 LRREESRGAHQRTDFPNRDDEHFLAHTLVHRESD---GTLRVGYLPVTIT
*.*:*:**.* * *:*:*** ::: **:.:::. . : :.: **. *
Mflv_4846|M.gilvum_PYR-GCK RYEPMERKY--
Mvan_1587|M.vanbaalenii_PYR-1 RYEPMERKY--
MSMEG_1670|M.smegmatis_MC2_155 RYEPMERKY--
MAB_3675|M.abscessus_ATCC_1997 RYEPMERKY--
TH_2182|M.thermoresistible__bu RYEPMERKY--
MLBr_00697|M.leprae_Br4923 RYEPQERKY--
MAV_4299|M.avium_104 RYEPKERKY--
MMAR_1201|M.marinum_M RYEPKERKY--
MUL_1369|M.ulcerans_Agy99 RYEPKERKY--
Rv1552|M.tuberculosis_H37Rv RWPPGERVYGR
Mb1578|M.bovis_AF2122/97 RWPPGERVYGR
*: * ** *