For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVRVGINGFGRIGRNFYRALLAQQEQGTADVEVVAANDITDNSTLAHLLKFDSILGRLPCDVGLEGDDT IVVGRAKIKALAVREGPAALPWGDLGVDVVVESTGLFTNAAKAKGHLDAGAKKVIISAPATDEDITIVLG VNDDKYDGSQNIISNASCTTNCLAPLAKVLDDEFGIVKGLMTTIHAYTQDQNLQDGPHKDLRRARAAALN IVPTSTGAAKAIGLVMPQLKGKLDGYALRVPIPTGSVTDLTVDLSTRASVDEINAAFKAAAEGRLKGILK YYDAPIVSSDIVTDPHSSIFDSGLTKVIDDQAKVVSWYDNEWGYSNRLVDLVTLVGKSL
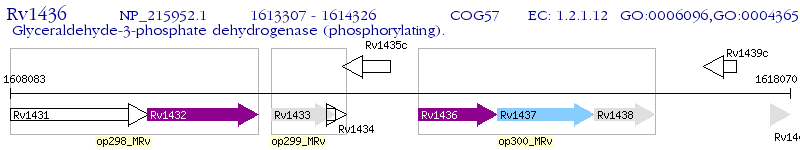
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1436 | gap | - | 100% (339) | glyceraldehyde-3-phosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1471 | gap | 0.0 | 100.00% (339) | glyceraldehyde-3-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3710 | - | 1e-166 | 85.00% (340) | glyceraldehyde-3-phosphate dehydrogenase, type I |
| M. leprae Br4923 | MLBr_00570 | gap | 1e-171 | 89.38% (339) | glyceraldehyde 3-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2779c | - | 1e-172 | 88.82% (340) | glyceraldehyde-3-phosphate dehydrogenase, type I |
| M. marinum M | MMAR_2239 | gap | 1e-179 | 92.04% (339) | glyceraldehyde 3-phosphate dehydrogenase Gap |
| M. avium 104 | MAV_3341 | gap | 0.0 | 94.10% (339) | glyceraldehyde-3-phosphate dehydrogenase, type I |
| M. smegmatis MC2 155 | MSMEG_3084 | gap | 1e-172 | 87.65% (340) | glyceraldehyde-3-phosphate dehydrogenase, type I |
| M. thermoresistible (build 8) | TH_0426 | gap | 1e-164 | 84.13% (334) | PROBABLE GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE GAP (GAPDH) |
| M. ulcerans Agy99 | MUL_1828 | gap | 1e-179 | 92.04% (339) | glyceraldehyde 3-phosphate dehydrogenase Gap |
| M. vanbaalenii PYR-1 | Mvan_2703 | - | 1e-168 | 86.47% (340) | glyceraldehyde-3-phosphate dehydrogenase, type I |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1436|M.tuberculosis_H37Rv ------------------------MTVRVGINGFGRIGRNFYRALLAQQE
Mb1471|M.bovis_AF2122/97 ------------------------MTVRVGINGFGRIGRNFYRALLAQQE
Mflv_3710|M.gilvum_PYR-GCK MERLLLGWLPSIADDSLIEPEETHVTIRVGVNGFGRIGRNFFRALDAQKA
Mvan_2703|M.vanbaalenii_PYR-1 ----------------MIEPEETDVTIRVGVNGFGRIGRNFFRALDAQKA
MAB_2779c|M.abscessus_ATCC_199 ------------------------MTVRVGVNGFGRIGRNFFRALDAQKA
MSMEG_3084|M.smegmatis_MC2_155 ------------------------MTIRVGVNGFGRIGRNFYRALATQKA
TH_0426|M.thermoresistible__bu ------------------------------VNGFGRIGRNFYRALAAQQA
MMAR_2239|M.marinum_M ------------------------MTVRVGINGFGRIGRNFYRALLAQQQ
MUL_1828|M.ulcerans_Agy99 ------------------------MTVRVGINGFGRIGRNFYRALLAQQQ
MAV_3341|M.avium_104 ------------------------MTVRVGINGFGRIGRNFYRALLAQQE
MLBr_00570|M.leprae_Br4923 ------------------------MTVRVGINGFGRIGRNFYRALLAQQE
:**********:*** :*:
Rv1436|M.tuberculosis_H37Rv QG-TADVEVVAANDITDNSTLAHLLKFDSILGRLPCDVGLEGDDTIVVGR
Mb1471|M.bovis_AF2122/97 QG-TADVEVVAANDITDNSTLAHLLKFDSILGRLPCDVGLEGDDTIVVGR
Mflv_3710|M.gilvum_PYR-GCK AGKNTDIEIIAVNDLTSNATLAHLLKFDSILGRLPYDVSLEGEDTIVVGD
Mvan_2703|M.vanbaalenii_PYR-1 DGKNTDIEIVAVNDLTDNATLAHLLKFDSILGRLPYDVSLEGEDTIVVGD
MAB_2779c|M.abscessus_ATCC_199 QGVNTDIEIVAVNDLTDNATLAHLLKFDSILGRLPYDVSLEGDDTIVVGD
MSMEG_3084|M.smegmatis_MC2_155 EGKNTDIEIVAVNDLTDNATLAHLLKFDSILGRLPQDVSLEGDDTIVIGD
TH_0426|M.thermoresistible__bu EGKSTDVEIVAVNDLTDNATLAHLLKFDSILGRLPEEVSLEGEDTIVIGS
MMAR_2239|M.marinum_M QG-SADIEVIAVNDITDNQTLAHLLKFDSILGRLPQDVSLEGEDTIVVGG
MUL_1828|M.ulcerans_Agy99 QG-SADIEVIAVNDITDNQTLAHLLKFDSILGRLPQDVSLEGEDTIVVGG
MAV_3341|M.avium_104 QG-TADIEVVAVNDITDNSTLAHLLKFDSILGRLPYDVSLEGEDTIVVGP
MLBr_00570|M.leprae_Br4923 HG-IADVQVVAINDITDNSTLAYLLKFDSILGRLPHDVSLEEEDTIVVGS
* :*::::* **:*.* ***:************ :*.** :****:*
Rv1436|M.tuberculosis_H37Rv AKIKALAVREGPAALPWGDLGVDVVVESTGLFTNAAKAKGHLDAGAKKVI
Mb1471|M.bovis_AF2122/97 AKIKALAVREGPAALPWGDLGVDVVVESTGLFTNAAKAKGHLDAGAKKVI
Mflv_3710|M.gilvum_PYR-GCK TKIKALEVKEGPAALPWGDLGVDVVVESTGIFTKRDKAQGHLDAGAKKVI
Mvan_2703|M.vanbaalenii_PYR-1 TKIKALEVKEGPAALPWGDLGVDVVVESTGIFTARAKAQGHLDAGAKKVI
MAB_2779c|M.abscessus_ATCC_199 HKIKALEVKEGPAALPWGDLGVDVVVESTGIFTARAKAQGHLDAGAKKVI
MSMEG_3084|M.smegmatis_MC2_155 TKIKALEVKEGPAALPWGDLGVDVVVESTGIFTNAAKAKGHLDAGAKKVI
TH_0426|M.thermoresistible__bu TKIKALAIKEGPSALPWGDLGVDVVLESTGIFTDAAKARGHLEAGAKKVI
MMAR_2239|M.marinum_M VKIKALEVREGPAAMPWGDLGVDVVVESTGLFTNAAKAKGHLDAGAKKVI
MUL_1828|M.ulcerans_Agy99 VKIKALEVREGPAAMPWGDLGVDVVVESTGLFTNAAKAKGHLDAGAKKVI
MAV_3341|M.avium_104 AKIKALEVREGPAALPWGDLGVDVVVESTGLFTNAAKAKGHLDAGAKKVI
MLBr_00570|M.leprae_Br4923 EKIKALAVREGPAALPWHAFGVDVVVESTGLFTNAAKAKGHLEAGAKKVI
***** ::***:*:** :*****:****:** **:***:*******
Rv1436|M.tuberculosis_H37Rv ISAPATDEDITIVLGVNDDKYDGSQNIISNASCTTNCLAPLAKVLDDEFG
Mb1471|M.bovis_AF2122/97 ISAPATDEDITIVLGVNDDKYDGSQNIISNASCTTNCLAPLAKVLDDEFG
Mflv_3710|M.gilvum_PYR-GCK ISAPASDEDITIVLGVNDDKYDGSQNIISNASCTTNCLGPLAKVLNDEFG
Mvan_2703|M.vanbaalenii_PYR-1 ISAPASDEDITIVLGVNDDKYDGSQNIISNASCTTNCLGPLAKVLNDEFG
MAB_2779c|M.abscessus_ATCC_199 ISAPASDEDITIVLGVNDDKYDGSQNIISNASCTTNCLGPLAKVLNDEFG
MSMEG_3084|M.smegmatis_MC2_155 ISAPATDEDITIVLGVNDDKYDGSQNIISNASCTTNCLGPLAKVLNDEFG
TH_0426|M.thermoresistible__bu ISAPAKGEDLTVVLGVNDDKYDGSQNIISNASCTTNCLAPLAKVLHDEFG
MMAR_2239|M.marinum_M ISAPATDPDLTIVLGVNDDKYDGSQNIISNASCTTNCLAPLAKVLHDEFG
MUL_1828|M.ulcerans_Agy99 ISAPATDPDLTIVLGVNDDKYDGSQNIISNASCTTNCLAPLAKVLHDEFG
MAV_3341|M.avium_104 ISAPATDEDITVVLGVNDDKYDGSQNIISNASCTTNCLAPLTKVLDDEFG
MLBr_00570|M.leprae_Br4923 VSAPATDPDITIVFGVNDDKYDGSQNIISNASCTTNCLAPLAKVLHDQFG
:****.. *:*:*:************************.**:***.*:**
Rv1436|M.tuberculosis_H37Rv IVKGLMTTIHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
Mb1471|M.bovis_AF2122/97 IVKGLMTTIHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
Mflv_3710|M.gilvum_PYR-GCK IVKGLMTTIHAYTQDQNLQDGPHGDLRRARAAAINIVPTSTGAAKAIGLV
Mvan_2703|M.vanbaalenii_PYR-1 IVKGLMTTVHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
MAB_2779c|M.abscessus_ATCC_199 IVKGLMTTIHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
MSMEG_3084|M.smegmatis_MC2_155 IVKGLMTTIHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
TH_0426|M.thermoresistible__bu IVKGLMTTVHAYTQDQNLQDGPHKDLRRARAAAINIVPTSTGAAKAIGLV
MMAR_2239|M.marinum_M IVKGLMTTIHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
MUL_1828|M.ulcerans_Agy99 IVKGLMTTIHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
MAV_3341|M.avium_104 IVRGLMTTIHAYTQDQNLQDGPHKDLRRARAAALNIVPTSTGAAKAIGLV
MLBr_00570|M.leprae_Br4923 IVKGLMTTVHAYTQDQNLQDGPHSDLRRARAAALNVVPTSTGAAKAIGLV
**:*****:************** *********:*:**************
Rv1436|M.tuberculosis_H37Rv MPQLKGKLDGYALRVPIPTGSVTDLTVDLSTRASVDEINAAFKAAAEGRL
Mb1471|M.bovis_AF2122/97 MPQLKGKLDGYALRVPIPTGSVTDLTVDLSTRASVDEINAAFKAAAEGRL
Mflv_3710|M.gilvum_PYR-GCK LPELKGKLDGYALRVPIPTGSVTDLTAELAKPGTVDEINAAMKAAADGPL
Mvan_2703|M.vanbaalenii_PYR-1 LPELKGKLDGYALRVPIPTGSCTDLTAELAKPGTAAEINAAMKAAAEGPL
MAB_2779c|M.abscessus_ATCC_199 LPELKGKLDGYALRVPIPTGSATDLTVELKKAASADEINAAMKAAAEGKL
MSMEG_3084|M.smegmatis_MC2_155 LPELKGKLDGYALRVPIPTGSVTDLTAELAKSASVEDINAAMKAAAEGPL
TH_0426|M.thermoresistible__bu MPELQGKLDGYALRVPIPTGSATDLTAELAKAATVDEINAAMKAAAEGPL
MMAR_2239|M.marinum_M MPELKGKLDGYALRVPIPTGSVTDLTAELAKAATAEEINAAFKAAAEGRL
MUL_1828|M.ulcerans_Agy99 MPELKGKLDGYALRVPIPTGSVTDLTAELAKAATAEEINAAFKAAAEGRL
MAV_3341|M.avium_104 MPNLKGKLDGYALRVPIPTGSVTDLTAELKKPASVEDINAAFKAAAEGRL
MLBr_00570|M.leprae_Br4923 MPELKGKLDGYALRVPIPTGSVTDLTADLSKCVSVNEINAVFQDAAEGRL
:*:*:**************** ****.:* . :. :***.:: **:* *
Rv1436|M.tuberculosis_H37Rv KGILKYYDAPIVSSDIVTDPHSSIFDSGLTKVIDDQAKVVSWYDNEWGYS
Mb1471|M.bovis_AF2122/97 KGILKYYDAPIVSSDIVTDPHSSIFDSGLTKVIDDQAKVVSWYDNEWGYS
Mflv_3710|M.gilvum_PYR-GCK KGILKYYDAPIVSSDIVTDPHSSLYDAGLTKVIDNQAKVVSWYDNEWGYS
Mvan_2703|M.vanbaalenii_PYR-1 KGILKYYDAPIVSSDIVTDPHSSIFDSGLTKVIDNQAKVVSWYDNEWGYS
MAB_2779c|M.abscessus_ATCC_199 KGILKYYDAPIVSSDIVTDPHSSIFDAGLTKVIDDQAKVVSWYDNEWGYS
MSMEG_3084|M.smegmatis_MC2_155 KGILKYYDAPIVSSDIVTDPHSSLYDAGLTKVIDNQAKVVSWYDNEWGYS
TH_0426|M.thermoresistible__bu KGILKYYDAPIVSSDIVTDPHSSLYDAGLTKVIDNQVKVVSWYDNEWGYA
MMAR_2239|M.marinum_M KGILKYYDAPIVSSDIVTDPHSSIFDSGLTKVIDDQAKVVSWYDNEWGYS
MUL_1828|M.ulcerans_Agy99 KGILKYYDAPIVSSDIVTDPHSSIFDSGLTKVIDDQAKVVSWYDNEWGYS
MAV_3341|M.avium_104 KGILKYYDAPIVSSDIVTDPHSSIFDSGLTKVIDNQAKVVSWYDNEWGYS
MLBr_00570|M.leprae_Br4923 KGILKYVDAPIVSSDIVTDPHSSIFDSGLTKVIASQAKVVSWYDNEWGYS
****** ****************::*:****** .*.************:
Rv1436|M.tuberculosis_H37Rv NRLVDLVTLVGKSL
Mb1471|M.bovis_AF2122/97 NRLVDLVTLVGKSL
Mflv_3710|M.gilvum_PYR-GCK NRLVDLVGLVGKSL
Mvan_2703|M.vanbaalenii_PYR-1 NRLVDLVGLVGKSL
MAB_2779c|M.abscessus_ATCC_199 NRLVDLVGLVGKSL
MSMEG_3084|M.smegmatis_MC2_155 NRLADLVALVGKSL
TH_0426|M.thermoresistible__bu NRCVDLVAMVGKSL
MMAR_2239|M.marinum_M NRLIDLVALVGKSL
MUL_1828|M.ulcerans_Agy99 NRLIDLVALVGKSL
MAV_3341|M.avium_104 NRLVDLVALVGKSL
MLBr_00570|M.leprae_Br4923 NRLVDLVGLVGKSL
** *** :*****