For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LGWTVKPGRVADGWQAPGVHLMARCSGPQPASERRADMDGGDIDAAVARVRAAGALAEPSRQPDDMSAEC ADDQGARCHLGQL*
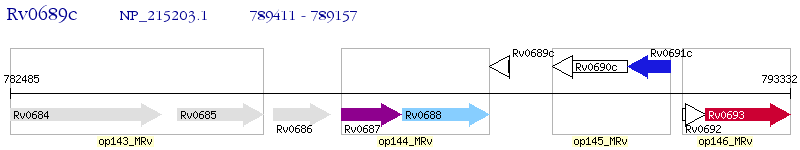
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0689c | - | - | 100% (84) | hypothetical protein Rv0689c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0708c | - | 9e-47 | 100.00% (84) | hypothetical protein Mb0708c |
| M. gilvum PYR-GCK | Mflv_5064 | - | 3e-16 | 50.59% (85) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1417 | - | 2e-15 | 45.88% (85) | glyoxalase family protein |
| M. thermoresistible (build 8) | TH_1887 | - | 2e-08 | 60.47% (43) | glyoxalase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1292 | - | 6e-15 | 45.35% (86) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5064|M.gilvum_PYR-GCK ---MTDPMDALRGGDLP-VDPDPAFARRLRSRLEAAANLFEQQPDRTKDI
Mvan_1292|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1887|M.thermoresistible__bu --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 MNADHDPLTVLHQHDEPPVAPDPEFVARLRARLEAALT----LPNRTEGV
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK IMSGTDTVLAELTEPT----SGGLIPYLTVTDARAAIDWYVDVLGASVVG
Mvan_1292|M.vanbaalenii_PYR-1 -MSGTDTALAELTRPAPIL-RSAVVPYLTVADARAAIRWYIDVFDAELVG
TH_1887|M.thermoresistible__bu --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 VMTGTDTAIAELKSPSQAAPRPAVISYLTVADARAAIGFYVDALGARVLG
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK APITMDDGRIGHAELTVSGATLYLADEYPEIGLKAPSSQHVSASMMLTVT
Mvan_1292|M.vanbaalenii_PYR-1 DPVEMDDGRIGHAELALADGVLYLADEYPEMGLKAPSPQAVSVSLMLTVA
TH_1887|M.thermoresistible__bu --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 DPIVMDDGRIGHAELLLAGGVLYLADEFPEIGLKAPAPNAVSVSLMLSVP
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK SVDGTLERARSARAFVQREPYEDHGARTAAIVDPFGHRWMLTGPITG--A
Mvan_1292|M.vanbaalenii_PYR-1 STDETLRRARDHGAQVQREPYENYGARNAALVDPFGHRWMLTGPVTG--A
TH_1887|M.thermoresistible__bu --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 DTDAALARAAAHGARVQREPYEDHGTRNATFIDPSGHRWMLSGPVGVRVP
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK PVPIRHGDVGYVSVRTPDAQRAAAFYGHVLGWVFDPETQRVTNTGQRIGI
Mvan_1292|M.vanbaalenii_PYR-1 AVPIQHGDVGYVAVWTPDAQRAATFYGHVLGWAVDPATQMVTNTGQRVGI
TH_1887|M.thermoresistible__bu --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 VEAIRHGDVGYVSVWVPDADRAAAFYGHVLGWTYDPATRQVTNTQEHIGI
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK TTAPGRHTLFCCYAVTDLDGARDSIVAGGGTVDDVVEFDFGSVLGATDPA
Mvan_1292|M.vanbaalenii_PYR-1 VSAPEQRTLFCCYAVTDLDGARASILAGGGSVGEVVDFGFGAVLRATDPH
TH_1887|M.thermoresistible__bu --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 FSVAGAPTMFCCYAVDDLDAARRAITAGGGTPGEVRESEFGTVLDATDPA
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK GAEFGVYRPAADEPRPLLNGGGPGELSYITYQVPDSAEFRAFYSRVLFWT
Mvan_1292|M.vanbaalenii_PYR-1 GVEFGVYRPASGEPRPLLNGGGHGELSYITYQVPDSTVFKAFYSGLLFWT
TH_1887|M.thermoresistible__bu --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 GTAFAVFEPVPGTPRPAVNGTGPGSLSYITYNVPDSAAFKEFYGHVLSWT
Rv0689c|M.tuberculosis_H37Rv ---------------------------------------------MLGWT
Mb0708c|M.bovis_AF2122/97 ---------------------------------------------MLGWT
Mflv_5064|M.gilvum_PYR-GCK YEPGRVDDGWGVVGAHPMSGAAGGHVEAVTVPMWTVADIDAAVARVREAG
Mvan_1292|M.vanbaalenii_PYR-1 YEPGRIDDGWGVVGAHPMSGAAGGNASAVTVPMWTVDDIDAAVSRVRSAG
TH_1887|M.thermoresistible__bu -----------------MAGVAGGIANQVTVPMWTVDDIDAAVQRVREAG
MSMEG_1417|M.smegmatis_MC2_155 FEPGRIADGWGVQNCQPMSGMAGGSDRATTVPMWTVADIDAAVTRVREAG
Rv0689c|M.tuberculosis_H37Rv VKPGRVADGWQAPGVHLMARCSGPQPASERRADMDGGDIDAAVARVRAAG
Mb0708c|M.bovis_AF2122/97 VKPGRVADGWQAPGVHLMARCSGPQPASERRADMDGGDIDAAVARVRAAG
*: :* . ****** *** **
Mflv_5064|M.gilvum_PYR-GCK GTVIEEPSQQPYGKSALCTDDQGARFYLGEF-----
Mvan_1292|M.vanbaalenii_PYR-1 GTVIEEPSQQSYGKSALCTDDQGTRFYLGEL-----
TH_1887|M.thermoresistible__bu GTVIEEPAQQPYGRSAQCLDDQGMRFYLGQFYLGQF
MSMEG_1417|M.smegmatis_MC2_155 GTVISAPARQPYGLMAECTDDQGARFYLGEF-----
Rv0689c|M.tuberculosis_H37Rv --ALAEPSRQPDDMSAECADDQGARCHLGQL-----
Mb0708c|M.bovis_AF2122/97 --ALAEPSRQPDDMSAECADDQGARCHLGQL-----
.: *::*. . * * **** * :**::