For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NADHDPLTVLHQHDEPPVAPDPEFVARLRARLEAALTLPNRTEGVVMTGTDTAIAELKSPSQAAPRPAVI SYLTVADARAAIGFYVDALGARVLGDPIVMDDGRIGHAELLLAGGVLYLADEFPEIGLKAPAPNAVSVSL MLSVPDTDAALARAAAHGARVQREPYEDHGTRNATFIDPSGHRWMLSGPVGVRVPVEAIRHGDVGYVSVW VPDADRAAAFYGHVLGWTYDPATRQVTNTQEHIGIFSVAGAPTMFCCYAVDDLDAARRAITAGGGTPGEV RESEFGTVLDATDPAGTAFAVFEPVPGTPRPAVNGTGPGSLSYITYNVPDSAAFKEFYGHVLSWTFEPGR IADGWGVQNCQPMSGMAGGSDRATTVPMWTVADIDAAVTRVREAGGTVISAPARQPYGLMAECTDDQGAR FYLGEF*
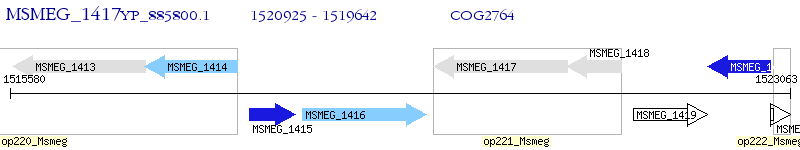
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1417 | - | - | 100% (427) | glyoxalase family protein |
| M. smegmatis MC2 155 | MSMEG_5680 | - | 1e-14 | 40.44% (136) | glyoxalase family protein |
| M. smegmatis MC2 155 | MSMEG_5616 | - | 5e-12 | 27.13% (247) | glyoxalase/bleomycin resistance protein/dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0708c | - | 1e-14 | 45.88% (85) | hypothetical protein Mb0708c |
| M. gilvum PYR-GCK | Mflv_5064 | - | 1e-152 | 63.47% (427) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv0689c | - | 1e-14 | 45.88% (85) | hypothetical protein Rv0689c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0931c | - | 3e-16 | 36.94% (157) | putative glyoxalase/bleomycin resistance protein |
| M. marinum M | MMAR_4645 | - | 1e-15 | 42.31% (130) | hypothetical protein MMAR_4645 |
| M. avium 104 | MAV_1014 | - | 8e-17 | 40.88% (137) | glyoxalase family protein |
| M. thermoresistible (build 8) | TH_1897 | - | 7e-94 | 63.96% (283) | glyoxalase family protein |
| M. ulcerans Agy99 | MUL_0263 | - | 6e-16 | 42.31% (130) | hypothetical protein MUL_0263 |
| M. vanbaalenii PYR-1 | Mvan_1292 | - | 1e-143 | 65.53% (380) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5064|M.gilvum_PYR-GCK -----MTDPMDALRGGDLPVDPDPAFARRLRSRLEAAANLFEQQPDRTKD
Mvan_1292|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 -----MNADHDPLTVLHQHDEPPVAPDPEFVARLRARLEAALTLPNRTEG
TH_1897|M.thermoresistible__bu MNQHHSRDPLDVLRGGDRPVQPDPAFAARLRARLESAARFLADRP-HTEG
MMAR_4645|M.marinum_M --------------------------------------------------
MUL_0263|M.ulcerans_Agy99 --------------------------------------------------
MAV_1014|M.avium_104 --------------------------------------------------
MAB_0931c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK IIMSGTDTVLAELT-------EPT----SGGLIPYLTVTDARAAIDWYVD
Mvan_1292|M.vanbaalenii_PYR-1 --MSGTDTALAELT-------RPAPIL-RSAVVPYLTVADARAAIRWYID
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 VVMTGTDTAIAELK-------SPSQAAPRPAVISYLTVADARAAIGFYVD
TH_1897|M.thermoresistible__bu VVMSGTQTALTELTGSTGTAEPAATTPPRPAAVPYLNVADARAAIDWYVE
MMAR_4645|M.marinum_M -------MAIN----------------VEPALSPHLVVHDAAAAIDFYVK
MUL_0263|M.ulcerans_Agy99 -------MAIN----------------VEPALSPHLVVHDAAAAIDFYVK
MAV_1014|M.avium_104 -------MAIN----------------IEPALSPHLVVDNAAAAIDFYVK
MAB_0931c|M.abscessus_ATCC_199 -------MEPSQTS--------PDLPPVQPSVSPYLIVEDSRAAIEFYKN
Mflv_5064|M.gilvum_PYR-GCK VLGASVVGAPITMDDGRIGHAELTVSGATLYLADEYPEIG----LKAPSS
Mvan_1292|M.vanbaalenii_PYR-1 VFDAELVGDPVEMDDGRIGHAELALADGVLYLADEYPEMG----LKAPSP
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 ALGARVLGDPIVMDDGRIGHAELLLAGGVLYLADEFPEIG----LKAPAP
TH_1897|M.thermoresistible__bu ALGATLSGEPVVMDDGRIGHAELTLAGGVFYLADEYPELG----LRAPAP
MMAR_4645|M.marinum_M AFGAEELG-RVPGPDGRLVHAALRINGSTVMLNDDFPEMCGGKSMTPVSL
MUL_0263|M.ulcerans_Agy99 AFGAEELG-RVPGPDGRLVHAALRINGSTVMLNDDFPEMCGGKSMTPVSL
MAV_1014|M.avium_104 AFGAEELG-RLPRPDGKLAHAAVRINGFMVMLNDDFPEVCGGKSMTPTSL
MAB_0931c|M.abscessus_ATCC_199 AFGAEELG-LLETPDGKVMHAAVKINGTTIMMNDDFPEYNDGKSSTPTAL
Mflv_5064|M.gilvum_PYR-GCK QHVSASMMLTVTSVDGTLERARSARAFVQREPYEDHGARTAAIVDPFGHR
Mvan_1292|M.vanbaalenii_PYR-1 QAVSVSLMLTVASTDETLRRARDHGAQVQREPYENYGARNAALVDPFGHR
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 NAVSVSLMLSVPDTDAALARAAAHGARVQREPYEDHGTRNATFIDPSGHR
TH_1897|M.thermoresistible__bu EAVSMSLVLPVADTDAALQRARRHGATVQREPYEAHGARSAAIVDPFGHR
MMAR_4645|M.marinum_M GGTPVTIHLTVTDVDAKFQ-------------------------------
MUL_0263|M.ulcerans_Agy99 DGTPVTIHLTVTDVDAKFQ-------------------------------
MAV_1014|M.avium_104 GGTPVTIHLTVPDVDASFQ-------------------------------
MAB_0931c|M.abscessus_ATCC_199 GGTPVTIHLTVPHVDDWFG-------------------------------
Mflv_5064|M.gilvum_PYR-GCK WMLTGPITG--APVPIRHGDVGYVSVRTPDAQRAAAFYGHVLGWVFDPET
Mvan_1292|M.vanbaalenii_PYR-1 WMLTGPVTG--AAVPIQHGDVGYVAVWTPDAQRAATFYGHVLGWAVDPAT
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 WMLSGPVGVRVPVEAIRHGDVGYVSVWVPDADRAAAFYGHVLGWTYDPAT
TH_1897|M.thermoresistible__bu WMLSGPLTG--ASARIQHGDIGYVALWVPDPQRAAQFYRQVLGWQWDPDS
MMAR_4645|M.marinum_M --------------------------------------------------
MUL_0263|M.ulcerans_Agy99 --------------------------------------------------
MAV_1014|M.avium_104 --------------------------------------------------
MAB_0931c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK QRVTNTGQRIGITTAPGRHTLFCCYAVTDLDGARDSIVAGGGTVDDVVEF
Mvan_1292|M.vanbaalenii_PYR-1 QMVTNTGQRVGIVSAPEQRTLFCCYAVTDLDGARASILAGGGSVGEVVDF
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 RQVTNTQEHIGIFSVAGAPTMFCCYAVDDLDAARRAITAGGGTPGEVRES
TH_1897|M.thermoresistible__bu NQVTNTTGPIGIHRVPDARTMFCCYAVTDLDAAR----------------
MMAR_4645|M.marinum_M --------------------------------------------------
MUL_0263|M.ulcerans_Agy99 --------------------------------------------------
MAV_1014|M.avium_104 --------------------------------------------------
MAB_0931c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK DFGSVLGATDPAGAEFGVYRPAADEPRPLLNGGGPGELSYITYQVPDSAE
Mvan_1292|M.vanbaalenii_PYR-1 GFGAVLRATDPHGVEFGVYRPASGEPRPLLNGGGHGELSYITYQVPDSTV
Mb0708c|M.bovis_AF2122/97 --------------------------------------------------
Rv0689c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1417|M.smegmatis_MC2_155 EFGTVLDATDPAGTAFAVFEPVPGTPRPAVNGTGPGSLSYITYNVPDSAA
TH_1897|M.thermoresistible__bu --------------------------------------------------
MMAR_4645|M.marinum_M --------------------------------------------------
MUL_0263|M.ulcerans_Agy99 --------------------------------------------------
MAV_1014|M.avium_104 --------------------------------------------------
MAB_0931c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_5064|M.gilvum_PYR-GCK FRAFYSRVLFWTYEPGRVDDGWGVVGAHPMSGAAGGHVEAVTVPMWTVAD
Mvan_1292|M.vanbaalenii_PYR-1 FKAFYSGLLFWTYEPGRIDDGWGVVGAHPMSGAAGGNASAVTVPMWTVDD
Mb0708c|M.bovis_AF2122/97 -------MLGWTVKPGRVADGWQAPGVHLMARCSGPQPASERRADMDGGD
Rv0689c|M.tuberculosis_H37Rv -------MLGWTVKPGRVADGWQAPGVHLMARCSGPQPASERRADMDGGD
MSMEG_1417|M.smegmatis_MC2_155 FKEFYGHVLSWTFEPGRIADGWGVQNCQPMSGMAGGSDRATTVPMWTVAD
TH_1897|M.thermoresistible__bu ---------------QAIIDGGGPPXXX-MVRHLLADPVLPDELLPAG--
MMAR_4645|M.marinum_M ----------------RALDAGATVVAPLDDQFWGDRYGVVADPFGHHWS
MUL_0263|M.ulcerans_Agy99 ----------------RALDAGATVVAPLDDQFWGERYGVVADPFGHHWS
MAV_1014|M.avium_104 ----------------RAVDAGATVVVPLEDQFWGDRYGMVADPFGHHWS
MAB_0931c|M.abscessus_ATCC_199 ----------------RAVDAGATVEMPLEDQFWGDRFGVIKDPFGHLWS
*.
Mflv_5064|M.gilvum_PYR-GCK IDAAVARVREAGGTVIEEPSQQPYGKSALCTDDQGARFYLGEF
Mvan_1292|M.vanbaalenii_PYR-1 IDAAVSRVRSAGGTVIEEPSQQSYGKSALCTDDQGTRFYLGEL
Mb0708c|M.bovis_AF2122/97 IDAAVARVRAAG--ALAEPSRQPDDMSAECADDQGARCHLGQL
Rv0689c|M.tuberculosis_H37Rv IDAAVARVRAAG--ALAEPSRQPDDMSAECADDQGARCHLGQL
MSMEG_1417|M.smegmatis_MC2_155 IDAAVTRVREAGGTVISAPARQPYGLMAECTDDQGARFYLGEF
TH_1897|M.thermoresistible__bu --WPGGAVRDAYRVFAAEMAARRDAGLVEAFD-----------
MMAR_4645|M.marinum_M LGQPVREVSMEEIQAAMSAQASN--------------------
MUL_0263|M.ulcerans_Agy99 LGQPVREVSMEEIQAAMSAQASN--------------------
MAV_1014|M.avium_104 LGQPVREVSPEEMAAAMAAMAAEQGAGQA--------------
MAB_0931c|M.abscessus_ATCC_199 LGQPVKIVNPEDLKKYAAGGSE---------------------
. *