For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPTIQQLVRKGRRDKISKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALRKVARVKLTSQVEVTAYIPGE GHNLQEHSMVLVRGGRVKDLPGVRYKIIRGSLDTQGVKNRKQARSRYGAKKEKG
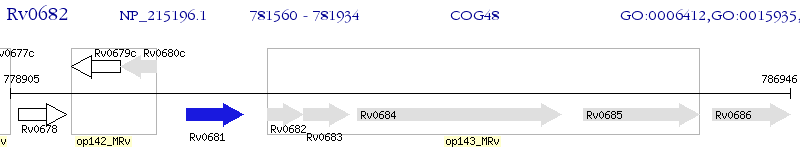
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0682 | rpsL | - | 100% (124) | 30S ribosomal protein S12 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0701 | rpsL | 3e-67 | 100.00% (124) | 30S ribosomal protein S12 |
| M. gilvum PYR-GCK | Mflv_5078 | rpsL | 2e-64 | 96.75% (123) | 30S ribosomal protein S12 |
| M. leprae Br4923 | MLBr_01880 | rpsL | 1e-65 | 97.56% (123) | 30S ribosomal protein S12 |
| M. abscessus ATCC 19977 | MAB_3851c | rpsL | 2e-64 | 96.75% (123) | 30S ribosomal protein S12 |
| M. marinum M | MMAR_1011 | rpsL | 7e-66 | 99.19% (123) | 30S ribosomal protein S12, RpsL |
| M. avium 104 | MAV_4492 | rpsL | 6e-66 | 99.19% (123) | 30S ribosomal protein S12 |
| M. smegmatis MC2 155 | MSMEG_1398 | rpsL | 5e-66 | 99.19% (123) | 30S ribosomal protein S12 |
| M. thermoresistible (build 8) | TH_1892 | rpsL | 2e-44 | 97.70% (87) | PROBABLE 30S RIBOSOMAL PROTEIN S12 RPSL |
| M. ulcerans Agy99 | MUL_0763 | rpsL | 4e-65 | 98.37% (123) | 30S ribosomal protein S12 |
| M. vanbaalenii PYR-1 | Mvan_1279 | rpsL | 2e-64 | 96.75% (123) | 30S ribosomal protein S12 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0682|M.tuberculosis_H37Rv MPTIQQLVRKGRRDKISKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
Mb0701|M.bovis_AF2122/97 MPTIQQLVRKGRRDKISKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
MMAR_1011|M.marinum_M MPTIQQLVRKGRRDKIGKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
MAV_4492|M.avium_104 MPTIQQLVRKGRRDKIGKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
MLBr_01880|M.leprae_Br4923 MPTIQQLVRKGRRDKIGKVKTAALKGNPQRRGVCTRVYTSTPKKPNSALR
MUL_0763|M.ulcerans_Agy99 MPTIQQLVRKGRRDKIGKVKTAALKRSPQRRGVCTRVYTTTPKKPNSALR
MSMEG_1398|M.smegmatis_MC2_155 MPTIQQLVRKGRRDKIAKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
TH_1892|M.thermoresistible__bu ------------------------------------VYTTTPKKPNSALR
Mflv_5078|M.gilvum_PYR-GCK MPTINQLVRKGRRDKIAKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
MAB_3851c|M.abscessus_ATCC_199 MPTINQLVRKGRRDKIAKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
Mvan_1279|M.vanbaalenii_PYR-1 MPTINQLVRKGRRDKIAKVKTAALKGSPQRRGVCTRVYTTTPKKPNSALR
***:**********
Rv0682|M.tuberculosis_H37Rv KVARVKLTSQVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
Mb0701|M.bovis_AF2122/97 KVARVKLTSQVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
MMAR_1011|M.marinum_M KVARVKLTSQVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
MAV_4492|M.avium_104 KVARVKLTSQVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
MLBr_01880|M.leprae_Br4923 KVARVKLTSQVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
MUL_0763|M.ulcerans_Agy99 KVARVKLTSQVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
MSMEG_1398|M.smegmatis_MC2_155 KVARVKLTSQVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
TH_1892|M.thermoresistible__bu KVARVKLTTGVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
Mflv_5078|M.gilvum_PYR-GCK KVARVRLTSAVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
MAB_3851c|M.abscessus_ATCC_199 KVARVRLTSAVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
Mvan_1279|M.vanbaalenii_PYR-1 KVARVRLTSAVEVTAYIPGEGHNLQEHSMVLVRGGRVKDLPGVRYKIIRG
*****:**: ****************************************
Rv0682|M.tuberculosis_H37Rv SLDTQGVKNRKQARSRYGAKKEKG
Mb0701|M.bovis_AF2122/97 SLDTQGVKNRKQARSRYGAKKEKG
MMAR_1011|M.marinum_M SLDTQGVKNRKQARSRYGAKKEKS
MAV_4492|M.avium_104 SLDTQGVKNRKQARSRYGAKKEKS
MLBr_01880|M.leprae_Br4923 SLDTQGVKNRKQARSRYGAKKEKS
MUL_0763|M.ulcerans_Agy99 SLDTQGVKNRKQARSRYGAKKEKS
MSMEG_1398|M.smegmatis_MC2_155 SLDTQGVKNRKQARSRYGAKKEKS
TH_1892|M.thermoresistible__bu SLDTQGVKNRKQARSRYGAKKEKS
Mflv_5078|M.gilvum_PYR-GCK SLDTQGVKNRKQARSRYGAKKEKS
MAB_3851c|M.abscessus_ATCC_199 SLDTQGVKNRKQARSRYGAKKEKS
Mvan_1279|M.vanbaalenii_PYR-1 SLDTQGVKNRKQARSRYGAKKEKS
***********************.