For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVESSTASAAAVLRARYPRTAASLDRYGGGTARRLERTGTFARFTRISVVQIGWALRRYRRETLRLVAEI GMGTGAMAVVGGTVAIIGFVTLSGGSLIAIQGFASLGNIGVEAFTGFFAALANTRVAAPIVSGVALAATV GAGATAQLGAMRISEEIDALEVMGIKSISFLVSTRILGGLVVIMPLYALALDMAFTSGQVVTTVFYGQSN GTYEHYFRTFLRPEDVGWSVVEVVIIAVVVMITHCYYGYTASGGPVGVGQAVGRSMRFSLVSVVVVVLLA ELALYGVDPNFNLTV
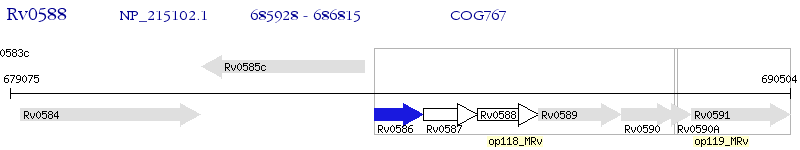
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0588 | yrbE2B | - | 100% (295) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE2B |
| M. tuberculosis H37Rv | Rv0168 | yrbE1B | e-123 | 76.39% (288) | integral membrane protein YRBE1B |
| M. tuberculosis H37Rv | Rv3500c | yrbE4B | 4e-79 | 51.67% (269) | integral membrane protein YrbE4b |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0603 | yrbE2B | 1e-161 | 100.00% (295) | integral membrane protein YrbE2b |
| M. gilvum PYR-GCK | Mflv_0712 | - | 1e-117 | 73.01% (289) | hypothetical protein Mflv_0712 |
| M. leprae Br4923 | MLBr_02588 | yrbE1B | 1e-121 | 76.04% (288) | hypothetical protein MLBr_02588 |
| M. abscessus ATCC 19977 | MAB_4154c | - | 2e-81 | 53.16% (269) | hypothetical protein MAB_4154c |
| M. marinum M | MMAR_0411 | yrbE1B | 1e-120 | 75.35% (288) | integral membrane protein YrbE1B |
| M. avium 104 | MAV_4552 | - | 1e-147 | 92.71% (288) | hypothetical protein MAV_4552 |
| M. smegmatis MC2 155 | MSMEG_0133 | - | 1e-112 | 70.49% (288) | ABC-transporter integral membrane protein |
| M. thermoresistible (build 8) | TH_4238 | - | 1e-117 | 73.10% (290) | PUTATIVE protein of unknown function DUF140 |
| M. ulcerans Agy99 | MUL_1061 | yrbE1B | 1e-119 | 74.65% (288) | integral membrane protein YrbE1B |
| M. vanbaalenii PYR-1 | Mvan_0133 | - | 1e-120 | 75.69% (288) | hypothetical protein Mvan_0133 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0588|M.tuberculosis_H37Rv -----MVESSTASAAAVLRARYPRTAASLDRYGGGTARRLERTGTFARFT
Mb0603|M.bovis_AF2122/97 -----MVESSTASAAAVLRARYPRTAASLDRYGGGTARRLERTGTFARFT
MAV_4552|M.avium_104 --------MAAISAPGALRARYPRTAANLDRYGGGTVRRLWQIGIFARFA
MMAR_0411|M.marinum_M -----------MSTAAVLRARFPRAIANFNRYAGSVTRGLDESGRLAWFI
MUL_1061|M.ulcerans_Agy99 -----------MSTAAVLRARFPRAIANFNRYAGSVTRGLDESGRLAWFI
MLBr_02588|M.leprae_Br4923 -----------MSTAAVLRARFPRAAANLNRYGGAAARGIDDIGQMSWFG
Mflv_0712|M.gilvum_PYR-GCK ----------MSDTAAVLRSRFPRGVARAEKIVGAPGRGLDSIGHVAWFV
Mvan_0133|M.vanbaalenii_PYR-1 ----------MS-TSAVLRSRFPRGVATAEKWAGAPARGLEAMGHVAVFV
TH_4238|M.thermoresistible__bu VFGSERGADMTVETGTVLRQRFPRAYSTGQRWVGAPARGLNSVGHVAWFV
MSMEG_0133|M.smegmatis_MC2_155 ----------MS-TVQVLRSRFPRAFSRSSEIAATPARFLDSMGHVAWFV
MAB_4154c|M.abscessus_ATCC_199 ------------MSGPYLSTHTVTSYT--VRYMRGLGRSFDKFGEQALFY
: * : : . * : * : *
Rv0588|M.tuberculosis_H37Rv RISVVQIGWALRRYRRETLRLVAEIGMGTGAMAVVGGTVAIIGFVTLSGG
Mb0603|M.bovis_AF2122/97 RISVVQIGWALRRYRRETLRLVAEIGMGTGAMAVVGGTVAIIGFVTLSGG
MAV_4552|M.avium_104 RISIGQIGWALRHYRRETLRLVAEIGMGTGAMAVVGGTVAIIGFVTLSGG
MMAR_0411|M.marinum_M LTSLGQSGHALRYYRKEILRLIAQIGMGTGAMAVVGGTAAIVGFVTLSGS
MUL_1061|M.ulcerans_Agy99 LTSLGQSGHALRYYRKEILRLIAQIGMGTGAMAVVGGTAAIVGFVTLSGS
MLBr_02588|M.leprae_Br4923 LIALGNIPNALSRYRKETLRLIAQIGMGTGAMAVVGGTAAIVGFVTLSGS
Mflv_0712|M.gilvum_PYR-GCK VTAVGSIGHAVRYYRKEMLRLIAEIGMGTGAMAVVGGTVAIVGFVTLSGS
Mvan_0133|M.vanbaalenii_PYR-1 VTAVGSIGHALRFYRREMLRLIAEIGMGTGAMAVVGGTVAIVGFVTLSGS
TH_4238|M.thermoresistible__bu VQAVGMIGHALRHYRRELLRLVAEIGMGTGAMAVVGGTVAIVGFVTLSGG
MSMEG_0133|M.smegmatis_MC2_155 VQAIVHVPHAFRHYRRESLRLVAEIGMGTGAMAVIGGTVAIIGFVTLSAG
MAB_4154c|M.abscessus_ATCC_199 AQSLSYIPAALTKYRKETIRVLAEITMGTGALVMIGGTVGVAVFLTLASG
:: *. **:* :*::*:* *****:.::***..: *:**:..
Rv0588|M.tuberculosis_H37Rv SLIAIQGFASLGNIGVEAFTGFFAALANTRVAAPIVSGVALAATVGAGAT
Mb0603|M.bovis_AF2122/97 SLIAIQGFASLGNIGVEAFTGFFAALANTRVAAPIVSGVALAATVGAGAT
MAV_4552|M.avium_104 SLIAIQGFASLGNIGVEAFTGFFAALANTRIAAPIVAGVTLAATVGAGAT
MMAR_0411|M.marinum_M SLIAIQGFASLGNIGVEAFTGFFSALVNVRIAGPVVTGVALAATVGAGAT
MUL_1061|M.ulcerans_Agy99 SLIAIQGFASLGNIGVEAFTGFFSALVNVRIAGPVVTGVALAATVGAGAT
MLBr_02588|M.leprae_Br4923 SLVAIQGFASLGAIGVEAFTGFFAALINVRIAAPVITGIVMAATVGAGAT
Mflv_0712|M.gilvum_PYR-GCK SLVAIQGFASLGNIGVEAFTGFFAALINVRIAAPVVAGQALAATVGAGAT
Mvan_0133|M.vanbaalenii_PYR-1 SLVAIQGFASLGNIGVEAFTGFFAALINVRIAAPVVAGQALAATVGAGAT
TH_4238|M.thermoresistible__bu SLIAIQGFASLGNIGVEAFTGFFAALVNVRIAAPVVAGQALAATVGAGAT
MSMEG_0133|M.smegmatis_MC2_155 SLIAIQGFASLGNIGVEAFTGFFAALANIRVVAPVVTGQALAATVGAGAT
MAB_4154c|M.abscessus_ATCC_199 GVIAVQGYSSLGNIGIEALTGFLSAFLNVRIVAPVVAGIAMAATIGAGTT
.::*:**::*** **:**:***::*: * *:..*:::* .:***:***:*
Rv0588|M.tuberculosis_H37Rv AQLGAMRISEEIDALEVMGIKSISFLVSTRILGGLVVIMPLYALALDMAF
Mb0603|M.bovis_AF2122/97 AQLGAMRISEEIDALEVMGIKSISFLVSTRILGGLVVIMPLYALALDMAF
MAV_4552|M.avium_104 AQLGAMRISEEIDALEVMGIKSISFLVSTRILGGLAVIVPLYALALDMAF
MMAR_0411|M.marinum_M AELGAMRISEEIDALEVMGIKSISYLASTRIMAGLVVIIPLYALAMIMSF
MUL_1061|M.ulcerans_Agy99 AELGAMRISEEIDALEVMGIQSISYLASTRIMAGLVVIIPLYALAMIMSF
MLBr_02588|M.leprae_Br4923 AELGAMRISEEIDALEVMSIKSISFLVTTRILAGLVVIVPLYAVAMIMAF
Mflv_0712|M.gilvum_PYR-GCK AELGAMRISEEIDALEVMGIKSISYLVSTRIVAGFVVIIPLYAMAIIMSF
Mvan_0133|M.vanbaalenii_PYR-1 AELGAMRISEEIDALEVMGIKSISYLVSTRIMAGFVVIIPLYAMAIIMSF
TH_4238|M.thermoresistible__bu AELGAMRISEEVDALEVMAIRSISYLVSTRIIAGFIVIIPLYAMAIIMSF
MSMEG_0133|M.smegmatis_MC2_155 AELGAMRISEEVDALEVMGIKSISYLVSTRIMAGAIVIIPLYAMAILLSF
MAB_4154c|M.abscessus_ATCC_199 AQLGAMRVAEEIDALETMAVHAISYLVSTRLVAGLIAIIPLYSLSVLAAF
*:*****::**:****.*.:::**:*.:**::.* .*:***:::: :*
Rv0588|M.tuberculosis_H37Rv TSGQVVTTVFYGQSNGTYEHYFRTFLRPEDVGWSVVEVVIIAVVVMITHC
Mb0603|M.bovis_AF2122/97 TSGQVVTTVFYGQSNGTYEHYFRTFLRPEDVGWSVVEVVIIAVVVMITHC
MAV_4552|M.avium_104 TSGQVVTTVFYGQSNGTYEHYFRTFLRPEDVGWSVFEVVIIAVVVMITHC
MMAR_0411|M.marinum_M FSPQMVTTVLYGQSSGTYEHYFRTFLRPDDVFWSFLEAIIIAAIVMVTHC
MUL_1061|M.ulcerans_Agy99 FSPQMVTTVLYGQSSGTYEHYFRTFLRPDDVFWSFLEAIIIAAIVMVTHC
MLBr_02588|M.leprae_Br4923 LSPHIITTVLYGQSNGTYEHYFRTFLRPDDVFWSFLEAVIITAVVMITHC
Mflv_0712|M.gilvum_PYR-GCK LSAQATTTIFYGQSTGTYEHYFRTFLRPDDVFWSFVQAVIISVIVMLNHC
Mvan_0133|M.vanbaalenii_PYR-1 LSAQVTTTVFYGQSVGTYEHYFRTFLRPDDVFWSFIQAVIISVIVMLNHC
TH_4238|M.thermoresistible__bu LSAQVTTTFFYGQSVGTYDHYFRTFLRPDDVMWSFVQAIIISVIVMLNHC
MSMEG_0133|M.smegmatis_MC2_155 MSAQLVTTIFYSQSVGTYEHYFHTFLRVDDVMWSFLEVIIMSVIVMLNHC
MAB_4154c|M.abscessus_ATCC_199 FASRATTVMINGQSPGVYDHYFNTFLVPTDLLWSFLQAIVMSVVVMLVHT
: : *..: .** *.*:***.*** *: **..:.::::.:**: *
Rv0588|M.tuberculosis_H37Rv YYGYTASGGPVGVGQAVGRSMRFSLVSVVVVVLLAELALYGVDPNFNLTV
Mb0603|M.bovis_AF2122/97 YYGYTASGGPVGVGQAVGRSMRFSLVSVVVVVLLAELALYGVDPNFNLTV
MAV_4552|M.avium_104 YYGYTASGGPVGVGHAVGRSMRFSLVSVVVVVLLAQLALYGVDPNFNLTV
MMAR_0411|M.marinum_M YYGYNAGGGPVGVGEAVGRSMRFSLVSVNVVVLCAALALYGVNPNFALTV
MUL_1061|M.ulcerans_Agy99 YYGYNAGGGPVGEGEAVGRSMRFSLVSVNVVVLCAALALYGVNPNFALTV
MLBr_02588|M.leprae_Br4923 YYGYTANGGPVGVGEAVGRSMRFSLVSVQVVVLSATLALYGVDPNFALTV
Mflv_0712|M.gilvum_PYR-GCK YYGFYASGGPVGVGEAVGRSMRASLVAIVCVVLFASLALYGVDPNFNLTV
Mvan_0133|M.vanbaalenii_PYR-1 YYGFYASGGPVGVGEAVGRSMRASLVAIVVVVLLASLALYGVDPNFNLTV
TH_4238|M.thermoresistible__bu YYGYYASGGPVGVGEAVGRSMRASLVAIVLVVLLASLALYGTDPNFNLTV
MSMEG_0133|M.smegmatis_MC2_155 YFGYFASGGAVGVGEAVGRSMRTSLIAIVLVVLLASLALYGTDPNFNLTV
MAB_4154c|M.abscessus_ATCC_199 SYGFNASGGPVGVGVAVGQAVRTSLIVVVLVTLFISLAVYGGSGNFNLSG
:*: *.**.** * ***:::* **: : *.* **:** . ** *: