For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VCGVRVAIVAESFLPQVNGVSNSVVKVLEHLRRTGHEALVIAPDTPPGEDRAERLHDGVRVHRVPSRMFP KVTTLPLGVPTFRMLRALRGFDPDVVHLASPALLGYGGLHAARRLGVPTVAVYQTDVPGFASSYGIPMTA RAAWAWFRHLHRLADRTLAPSTATMESLIAQGIPRVHRWARGVDVQRFAPSARNEVLRRRWSPDGKPIVG FVGRLAPEKHVDRLTGLAASGAVRLVIVGDGIDRARLQSAMPTAVFTGARYGKELAEAYASMDVFVHSGE HETFCQVVQEALASGLPVIAPDAGGPRDLITPHRTGLLLPVGEFEHRLPDAVAHLVHERQRYALAARRSV LGRSWPVVCDELLGHYEAVRGRRTTQAA
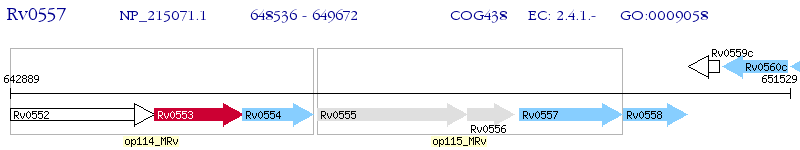
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0557 | pimB | - | 100% (378) | MANNOSYLTRANSFERASE PIMB |
| M. tuberculosis H37Rv | Rv2188c | - | 3e-12 | 26.98% (378) | hypothetical protein Rv2188c |
| M. tuberculosis H37Rv | Rv2610c | pimA | 4e-12 | 26.94% (386) | alpha-mannosyltransferase PIMA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0572 | pimB | 0.0 | 100.00% (378) | mannosyltransferase PIMB |
| M. gilvum PYR-GCK | Mflv_5260 | - | 1e-168 | 76.80% (375) | glycosyl transferase, group 1 |
| M. leprae Br4923 | MLBr_00886 | - | 7e-15 | 27.27% (374) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_3931c | - | 1e-143 | 67.84% (370) | glycosyl transferase |
| M. marinum M | MMAR_0903 | pimB | 1e-180 | 84.43% (366) | mannosyltransferase, PimB |
| M. avium 104 | MAV_4586 | - | 0.0 | 86.61% (366) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_1113 | - | 1e-165 | 74.93% (375) | glycosyl transferase |
| M. thermoresistible (build 8) | TH_1121 | - | 1e-165 | 77.32% (366) | MANNOSYLTRANSFERASE PIMB |
| M. ulcerans Agy99 | MUL_0656 | pimB | 1e-177 | 83.33% (366) | mannosyltransferase, PimB |
| M. vanbaalenii PYR-1 | Mvan_0978 | - | 1e-166 | 74.67% (375) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5260|M.gilvum_PYR-GCK ---MRVAIVAESFLPNVNGVTNSVLRVIEHLRRTG-HEALVIAPDTPRGE
Mvan_0978|M.vanbaalenii_PYR-1 ---MRVAIVAESFLPNVNGVTNSVLRVIEHLRRTG-HEVLVIAPDTPRGQ
MSMEG_1113|M.smegmatis_MC2_155 ---MRVAIVAESFLPNVNGVTNSVLRVIDHLRRTG-HEVLVIAPDTPRGQ
TH_1121|M.thermoresistible__bu ---VRVAIVTESFLPHVNGVTNSVLRVVEHLRGTG-HEVLVIAPDTPPGQ
Rv0557|M.tuberculosis_H37Rv MCGVRVAIVAESFLPQVNGVSNSVVKVLEHLRRTG-HEALVIAPDTPPGE
Mb0572|M.bovis_AF2122/97 MCGVRVAIVAESFLPQVNGVSNSVVKVLEHLRRTG-HEALVIAPDTPPGE
MAV_4586|M.avium_104 ---MRVAIVAESFLPEVNGVSNSVIRVLEHLRRTG-HEALVIAPDTPPGQ
MMAR_0903|M.marinum_M ---MRVAIVAESFLPNVNGVSNSVVRVLEHLRRTG-HEALVIAPDNPPGE
MUL_0656|M.ulcerans_Agy99 ---MRVAIVAESFLPNVNGVSNSVVRVLEHLRRTG-HEALVIAPDNPPGE
MAB_3931c|M.abscessus_ATCC_199 ---MRIAIVAESFLPNVNGVSNSVLRVLEHFRRTG-HEAIVIAPDTPRGQ
MLBr_00886|M.leprae_Br4923 --MSRVLLVTNDFPPRRGGIQSYLGEFVDRLINIGSHDVMVYAPQWTGAD
*: :*::.* *. .*: . : ..:::: * *:.:* **: . .:
Mflv_5260|M.gilvum_PYR-GCK PPADRVYDGVRVHRVPSAMFPKVTSLPLGVPRPRMVGVLRGFDPDVVHLA
Mvan_0978|M.vanbaalenii_PYR-1 PAADKVYEGVRVHRVPSMMFPKITSLPLGVPRPRMVGVLRGFDPDVVHLA
MSMEG_1113|M.smegmatis_MC2_155 PPADRIHDGVRVHRVPSRMFPKITSLPLGVPRPRMIGVLRGFDPDVVHLA
TH_1121|M.thermoresistible__bu PPAERVHDGVRIHRVPSRMFPKITSLPLGIPRPRMIGVLRGFDPDVVHLA
Rv0557|M.tuberculosis_H37Rv DRAERLHDGVRVHRVPSRMFPKVTTLPLGVPTFRMLRALRGFDPDVVHLA
Mb0572|M.bovis_AF2122/97 DRAERLHDGVRVHRVPSRMFPKVTTLPLGVPTFRMLRALRGFDPDVVHLA
MAV_4586|M.avium_104 PRAERIHDGIRVHRVPSRMFPKVTTLPLGVPTPRLVSVLRGFDPHVVHLA
MMAR_0903|M.marinum_M PRADRLHDGVRVHRTPARMFPKVTTLPLGVPTPRILRVLRGFEPDVVHLA
MUL_0656|M.ulcerans_Agy99 PRADRLHDGVRVHRTPARMFPEVTTLPLGVPTPRILRVLRGFEPDVVHLA
MAB_3931c|M.abscessus_ATCC_199 APAATEYAGVPVHRVPAIMFPKVSTLPLGVPQPRMVRIFREFAPDVVHLA
MLBr_00886|M.leprae_Br4923 TFDNAAHDGG----YQVVRHPGTLMLPGPAVDARMRRLIVDHGIDTVWFG
: * .* ** *: : . ..* :.
Mflv_5260|M.gilvum_PYR-GCK SPALLGWGGVHAARHLGVPTVAVFQTDVAGFAQSYGIGMMSRASWAWTRR
Mvan_0978|M.vanbaalenii_PYR-1 SPALLGWGGVHAARRLGIPTVAVFQTDVAGFAESYGIGMLSHASWAWTRR
MSMEG_1113|M.smegmatis_MC2_155 SPALLGYGGLHAARHLGVPSVAVFQTDVAGFAESYGMGVASRAAWAWTRH
TH_1121|M.thermoresistible__bu SPALLGYGGLLAARRLRVPTVAVFQTDVAGFAASYGMGVAARAAWAWTRH
Rv0557|M.tuberculosis_H37Rv SPALLGYGGLHAARRLGVPTVAVYQTDVPGFASSYGIPMTARAAWAWFRH
Mb0572|M.bovis_AF2122/97 SPALLGYGGLHAARRLGVPTVAVYQTDVPGFASSYGIPMTARAAWAWFRH
MAV_4586|M.avium_104 SPALLGYGGLRAARWLGVPTVAVYQTDVPGFAASYGIPMTARAAWAWFRH
MMAR_0903|M.marinum_M SPALLGYGALRAARRLGVPTVAVYQTDVPGFAASYGIAATSRAAWAWFRH
MUL_0656|M.ulcerans_Agy99 SPALLGYGALRAARRLGVPTVAVYQTDVLGFAASYGIAATSRAAWAWFRH
MAB_3931c|M.abscessus_ATCC_199 SPFVLGYGGLRAAQHLDVPVVAVYQTDVAGFADSYGAGFAARAAWRWTKH
MLBr_00886|M.leprae_Br4923 SAAPLALLAPCARKAGATRVLASTHGHEVGWSMLPVARSVLRRIGDVTDV
*. *. . * : :* : . *:: :
Mflv_5260|M.gilvum_PYR-GCK LHSKADRTLAPSTSAMENLAAHGIPRVHKWARGVDVTGFAP-SARDQRLR
Mvan_0978|M.vanbaalenii_PYR-1 LHSKADRTLAPSTSAMENLAAHRIPRVHKWARGVDVTGFAP-SARDERLR
MSMEG_1113|M.smegmatis_MC2_155 LHSRADRTLAPSTSAMENLAAHRIPRVHRWGRGVDITGFVP-SARDEHLR
TH_1121|M.thermoresistible__bu LHRLADRTLAPSTAAMRDLARHGVPRVHLWARGVDVGGFAP-SARDEQLR
Rv0557|M.tuberculosis_H37Rv LHRLADRTLAPSTATMESLIAQGIPRVHRWARGVDVQRFAP-SARNEVLR
Mb0572|M.bovis_AF2122/97 LHRLADRTLAPSTATMESLIAQGIPRVHRWARGVDVQRFAP-SARNEVLR
MAV_4586|M.avium_104 LHTLADRTLAPSTVTMESLVAHRFPRVHRWARGVDVLRFAP-SARSEALR
MMAR_0903|M.marinum_M LHGLADRTLAPSTPTMQTLLDQGFPRVHRWARGVDLIRFAP-SARDESLR
MUL_0656|M.ulcerans_Agy99 LHGLADRTLAPSTPTMQTLLDQGFPRVHRWARGVDLIRFAP-SARDESLR
MAB_3931c|M.abscessus_ATCC_199 LHGRADRTLAPSSSAMEDLAMHGIPRVFRWSRGVDIERFAP-SARDDELR
MLBr_00886|M.leprae_Br4923 VTFVSRYTRSRVTAAFGPRAALEY-----LPPGVDADRFRPDPASRAELR
: : * : : :: *** * * .* **
Mflv_5260|M.gilvum_PYR-GCK QSWSPQGKPIVGFVGRLAPEKH----VERLAPLHARDD-IQLVIVGDGVD
Mvan_0978|M.vanbaalenii_PYR-1 RSWSPDGRPIIGFVGRLAPEKH----VERLAALHARDD-LQIVVVGDGVD
MSMEG_1113|M.smegmatis_MC2_155 RTWSPDGRPIVGFVGRLAPEKH----VERLAVLAARDD-LQLVIVGDGVD
TH_1121|M.thermoresistible__bu RRWSPDGKPIVGFVGRLAPEKH----VERLRALAPRTD-LQLVIVGDGVD
Rv0557|M.tuberculosis_H37Rv RRWSPDGKPIVGFVGRLAPEKH----VDRLTGLAASGA-VRLVIVGDGID
Mb0572|M.bovis_AF2122/97 RRWSPDGKPIVGFVGRLAPEKH----VDRLTGLAASGA-VRLVIVGDGID
MAV_4586|M.avium_104 RRWSPQGKPIVGFVGRLAPEKH----VERLAGLAASDA-VQLVVVGDGVD
MMAR_0903|M.marinum_M RRWSPDGRPIVGFVGRLAPEKH----AERLAGLAASGA-VTLVIVGAGVD
MUL_0656|M.ulcerans_Agy99 RRWSPDGRPIVGFVGRLAPEKD----AERLAGLAASGA-VTLVIVGAGVD
MAB_3931c|M.abscessus_ATCC_199 RAWSPKGKPIIGFVGRLAPEKH----VERLAVLAGRDD-LQLVIVGGGVE
MLBr_00886|M.leprae_Br4923 NRYRLGERPTVVCLARLVPRKGQDMLIRALPSIRQRADGAALMIVGGGPY
. : :* : :.**.*.* * : :::** *
Mflv_5260|M.gilvum_PYR-GCK RARLESAFP------RAVFTGALYGPELAAAYASMDVFVHGG-------E
Mvan_0978|M.vanbaalenii_PYR-1 RAKLESELP------RAVFTGALYGDELAAAYASMDVFVHPG-------E
MSMEG_1113|M.smegmatis_MC2_155 RVKLQTVLP------TAVFTGELRGAALAAAYASMDVFVHPG-------E
TH_1121|M.thermoresistible__bu RRKLENLLP------TAVFTGALYGEQLAAAYASMDVFVHPG-------E
Rv0557|M.tuberculosis_H37Rv RARLQSAMP------TAVFTGARYGKELAEAYASMDVFVHSG-------E
Mb0572|M.bovis_AF2122/97 RARLQSAMP------TAVFTGARYGKELAEAYASMDVFVHSG-------E
MAV_4586|M.avium_104 RAKLQSAMP------TAVFTGALYGDELAAAYASMDVFVHPG-------E
MMAR_0903|M.marinum_M RRKLELAMP------TAVFTGALYGDELAAAYASMDVFVHAG-------E
MUL_0656|M.ulcerans_Agy99 RRKLELAMP------TAVFTGALYGDELAAAYASMDVFVHAG-------E
MAB_3931c|M.abscessus_ATCC_199 REALEKLMP------TAVFTGELTGSALSRAYASLDVFIHPG-------E
MLBr_00886|M.leprae_Br4923 LGTLRKLAQGCGVADHVIFAGGVAAVEVPAHYTLADVFAMPCRTRGGGMD
*. .:*:* . :. *: *** :
Mflv_5260|M.gilvum_PYR-GCK HETFCQAVQEAMASGLPVIAPDAGGPRDLVAPYRSGLLLPVAEFEDRLSE
Mvan_0978|M.vanbaalenii_PYR-1 HETFCQAVQEAMASGLPVIAPDAGGPRDLVAPYRTGLLLPVAEFESRLAE
MSMEG_1113|M.smegmatis_MC2_155 HETFCQTVQEAMASGVPVIAPDAGGPRDLVAPCRTGLLLDVDGFECALPA
TH_1121|M.thermoresistible__bu HETFCQAVQEALASGLPVIAPDAGGPRDLVAPCRTGLLLPVEEFETRLAG
Rv0557|M.tuberculosis_H37Rv HETFCQVVQEALASGLPVIAPDAGGPRDLITPHRTGLLLPVGEFEHRLPD
Mb0572|M.bovis_AF2122/97 HETFCQVVQEALASGLPVIAPDAGGPRDLITPHRTGLLLPVGEFEHRLPD
MAV_4586|M.avium_104 HETFCQVVQEALASGLPVIAPDAGGPRDLVTPWRTGLLLAVDEFETKLPA
MMAR_0903|M.marinum_M HETFCQVVQEALASGLPVIAPDAGGPRDLVTPQRTGLLLPVDGFETQLPA
MUL_0656|M.ulcerans_Agy99 HETFCQVVQEALASGLPVIAPDAGGPRDLVAPQRTGLLLPVDGFETQLPA
MAB_3931c|M.abscessus_ATCC_199 HETFCQAVQEALASGVPSIAPDAGGPRDLVIPGRNGMLLPVADFEARLGM
MLBr_00886|M.leprae_Br4923 VEGLGIVFLEASATGVPVIAGKSGGAPEAVQHNKTGLVVDGRSVN-MVAD
* : .. ** *:*:* ** .:**. : : :.*::: .: :
Mflv_5260|M.gilvum_PYR-GCK SVDHLVV--ERRRYSQAARRSVLGRTWPVICDELVGHYEAVVG---ARAL
Mvan_0978|M.vanbaalenii_PYR-1 SVDHLVA--ERRRYSVAARRSVLGRTWPVICDELLGHYEAVMG---ARRL
MSMEG_1113|M.smegmatis_MC2_155 AVTHLIA--ERRRYGIAARRSVLARTWPVVCDELIGHYEAVLG---RRSL
TH_1121|M.thermoresistible__bu AVDHLLA--ERPRYAVAARRSVLGRTWPAICDQLLEHYAAVXX---AVAH
Rv0557|M.tuberculosis_H37Rv AVAHLVH--ERQRYALAARRSVLGRSWPVVCDELLGHYEAVRG---RRTT
Mb0572|M.bovis_AF2122/97 AVAHLVH--ERQRYALAARRSVLGRSWPVVCDELLGHYEAVRG---RRTT
MAV_4586|M.avium_104 AVAHLIG--ERHRYAPAARRSVLGRSWPVICDELLGHYEAVLSPFVRRQV
MMAR_0903|M.marinum_M AVAHLVH--ERQRYSQAARRSVLGRSWSVICDELLDHYAEVLAPTGRAQD
MUL_0656|M.ulcerans_Agy99 AVAHLVH--ERQRYSQAARRSVLGRSWSVICDELLDHYAEVLAPTGRAQD
MAB_3931c|M.abscessus_ATCC_199 AVDHLVQPATRSRFSAAARRGVLARTWPAICDELLEHYAAAAGTTLGRGI
MLBr_00886|M.leprae_Br4923 AVTELLTDRDRAAAMGAAGRHWVTAEWR--WDTLAARLADLLCGNDSR--
:* .*: * ** * : * * * :
Mflv_5260|M.gilvum_PYR-GCK RAA------
Mvan_0978|M.vanbaalenii_PYR-1 RAA------
MSMEG_1113|M.smegmatis_MC2_155 RAA------
TH_1121|M.thermoresistible__bu PAPHRHAG-
Rv0557|M.tuberculosis_H37Rv QAA------
Mb0572|M.bovis_AF2122/97 QAA------
MAV_4586|M.avium_104 RAGRYAQGE
MMAR_0903|M.marinum_M WLWRRRRA-
MUL_0656|M.ulcerans_Agy99 WLWRRRRA-
MAB_3931c|M.abscessus_ATCC_199 MTA------
MLBr_00886|M.leprae_Br4923 ---------