For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLFGMARIAIIGGGSIGEALLSGLLRAGRQVKDLVVAERMPDRANYLAQTYSVLVTSAADAVENATFVVV AVKPADVEPVIADLANATAAAENDSAEQVFVTVVAGITIAYFESKLPAGTPVVRAMPNAAALVGAGVTAL AKGRFVTPQQLEEVSALFDAVGGVLTVPESQLDAVTAVSGSGPAYFFLLVEALVDAGVGVGLSRQVATDL AAQTMAGSAAMLLERMEQDQGGANGELMGLRVDLTASRLRAAVTSPGGTTAAALRELERGGFRMAVDAAV QAAKSRSEQLRITPE
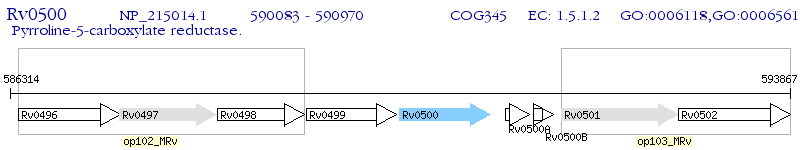
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0500 | proC | - | 100% (295) | pyrroline-5-carboxylate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0511 | proC | 1e-157 | 99.66% (295) | pyrroline-5-carboxylate reductase |
| M. gilvum PYR-GCK | Mflv_0075 | - | 1e-112 | 72.20% (295) | pyrroline-5-carboxylate reductase |
| M. leprae Br4923 | MLBr_02430 | proC | 1e-127 | 82.37% (295) | pyrroline-5-carboxylate reductase |
| M. abscessus ATCC 19977 | MAB_4005c | - | 1e-99 | 64.55% (299) | pyrroline-5-carboxylate reductase |
| M. marinum M | MMAR_0826 | proC | 1e-126 | 81.69% (295) | pyrroline-5-carboxylate reductase ProC |
| M. avium 104 | MAV_4652 | proC | 1e-127 | 81.02% (295) | pyrroline-5-carboxylate reductase |
| M. smegmatis MC2 155 | MSMEG_0943 | proC | 1e-110 | 72.51% (291) | pyrroline-5-carboxylate reductase |
| M. thermoresistible (build 8) | TH_0955 | proC | 1e-106 | 69.42% (291) | PROBABLE PYRROLINE-5-CARBOXYLATE REDUCTASE PROC (P5CR) (P5C |
| M. ulcerans Agy99 | MUL_4570 | proC | 1e-125 | 81.36% (295) | pyrroline-5-carboxylate reductase |
| M. vanbaalenii PYR-1 | Mvan_0837 | - | 1e-111 | 71.86% (295) | pyrroline-5-carboxylate reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0075|M.gilvum_PYR-GCK ------------MLARMARIAIIGGGSIGEALLAGLLRAGRQVKDLVVAE
Mvan_0837|M.vanbaalenii_PYR-1 MSRQVCAHRERAMLCGMARIAIIGGGSIGEALLAGLLRAGRQVKDLVVAE
MSMEG_0943|M.smegmatis_MC2_155 ----------------MSRIAIIGGGSMGEALLSGLLRAGRQVKDMVVAE
TH_0955|M.thermoresistible__bu ----------------VARIAIIGGGNIGEALLSGLLRAGRQVKDLVVAE
Rv0500|M.tuberculosis_H37Rv ------------MLFGMARIAIIGGGSIGEALLSGLLRAGRQVKDLVVAE
Mb0511|M.bovis_AF2122/97 ------------MLFGMARIAIIGGGSIGEALLSGLLRAGRQVKDLVVAE
MLBr_02430|M.leprae_Br4923 ------------MLSSMARIAIIGGGSIGEALLSGLLRAGRQVKDLVVAE
MMAR_0826|M.marinum_M ------------MLSGMPRIAIIGGGSIGEALLSGLLRAGRQVKDLVVAE
MUL_4570|M.ulcerans_Agy99 ------------MLSGMPRIAIIGGGSIGEALLSGLLRAGRQVKDLVVAE
MAV_4652|M.avium_104 ------------MLSGMARIAIIGGGSIGEALLSGLLRAGRQVKDLVVVE
MAB_4005c|M.abscessus_ATCC_199 ------------MLVAMSRIAIIGGGNIGEALISGLLRAGRQAKDIVVSE
:.********.:****::********.**:** *
Mflv_0075|M.gilvum_PYR-GCK KSPDRGKYLAEKYSVLVTTVADAVDAATYVIVAVKPADVQHVIGDIADTA
Mvan_0837|M.vanbaalenii_PYR-1 KNPGRAKYLAEKYSVLVTAVSDAVDAATYVVVAVKPSDVQYVIGDIADTA
MSMEG_0943|M.smegmatis_MC2_155 KFPERAKYLADKYSVRVTSVADAAENAHYVVVAVKPADVEKVIGEIAEAA
TH_0955|M.thermoresistible__bu KDPNRAKYLAEKYGVLVTSVGDAAESATFIVIAVKPGDVEKVIDDIAEAA
Rv0500|M.tuberculosis_H37Rv RMPDRANYLAQTYSVLVTSAADAVENATFVVVAVKPADVEPVIADLANAT
Mb0511|M.bovis_AF2122/97 RMPDRANYLAQTYSVLVTSAADAVENATFVVVAVKPADVEPVIADLANAT
MLBr_02430|M.leprae_Br4923 RMPDRARYLADTYSVLVTSVTDAVENAMFVVVAVKPTDVESVMGDLVQAA
MMAR_0826|M.marinum_M RVPERAKYLASTYSVLVTSAKDAAESASLVVVAVKPGDVDSVMGDLSAVV
MUL_4570|M.ulcerans_Agy99 RVPERAKYLASTYSVLVTSAKDAAESASLVVVAVKPGDVDSVMGDLSAVV
MAV_4652|M.avium_104 RVPERAKYLADTYSVLMTSVADAVENASFVVVAVKPADVESVMSEIARAA
MAB_4005c|M.abscessus_ATCC_199 KVPARARALADAYSIRISEVADAVEGADFIVVAVKPSDVESATTEIAAAL
: * *.. **. *.: :: . **.: * :::**** **: . :: .
Mflv_0075|M.gilvum_PYR-GCK ARADSDAAE----QVFVSVAAGVSTSFYESKLPAGSPVIRVMPNAPMLVG
Mvan_0837|M.vanbaalenii_PYR-1 ARAESNAAE----QVFVTVAAGVSTAYYENKLPAGSPVVRVMPNAPMLVG
MSMEG_0943|M.smegmatis_MC2_155 ANAETDTAE----QVFVTIAAGVGTAYYEAKLPAGSPVIRVMPNAPVIVG
TH_0955|M.thermoresistible__bu ARADSGTDE----QVFVSVAAGVGTGYYEAKLPAGAPVVRVMPNAPMVVG
Rv0500|M.tuberculosis_H37Rv AAAENDSAE----QVFVTVVAGITIAYFESKLPAGTPVVRAMPNAAALVG
Mb0511|M.bovis_AF2122/97 AAAENDSAE----QVFVTVVAGITIAYFESKLPAGTPVVRAMPNAAALVG
MLBr_02430|M.leprae_Br4923 AVA-NDSAE----QVLVTVAAGVTITYLESKLPAGTPVVRAMPNAAALVG
MMAR_0826|M.marinum_M AAAESYSGE----QVFVTVAAGVTTSYFESKLPAGTPVVRAMPNAAALVS
MUL_4570|M.ulcerans_Agy99 AAAESYSGE----QVFVTVAAGVTTSYFESKLPAGTPVVRAMPNAAALVS
MAV_4652|M.avium_104 GQAESDTAE----QVFVTVAAGVTIGYFESRLPAGTPVVRAMPNAAALVG
MAB_4005c|M.abscessus_ATCC_199 AKLDAEGSERETEQVLVSVAAGVSTGFFESKLAAGAPVVRVMPNAPMLVG
. * **:*::.**: : * :*.**:**:*.****. :*.
Mflv_0075|M.gilvum_PYR-GCK GGVSALAPGRFASPEQLKEVSLLFDAVGGVLTVAESQIDAVTAVSGSGPA
Mvan_0837|M.vanbaalenii_PYR-1 GGVSALAPGRFATQEHLKEVSLLFDAVGGVLTVAESQIDAVTAVSGSGPA
MSMEG_0943|M.smegmatis_MC2_155 GGVSALAPGRFATPEQLKEVSTLFDAVGGVITVSESQLDAVTALSGSGPA
TH_0955|M.thermoresistible__bu GGVSALAPGRFATPEQLKEVAGIFDAVGGVLTVTEAQMDAVTAVSGSGPA
Rv0500|M.tuberculosis_H37Rv AGVTALAKGRFVTPQQLEEVSALFDAVGGVLTVPESQLDAVTAVSGSGPA
Mb0511|M.bovis_AF2122/97 AGVTALAKGRFVTPQQLEEVSALFDAVGGVLTVPESQLDAVTAVSGSGPA
MLBr_02430|M.leprae_Br4923 AGVTVLAKGRFVTGQQFEDVLAMFDAVGGVLTVPESQMDAVTAVSGSGPA
MMAR_0826|M.marinum_M AGVTALAKGRFVTPEQLEEVSALFDSVGVVLTVPEQQLDAVTALSGSGPA
MUL_4570|M.ulcerans_Agy99 AGVTALAKGRFVTPEQLEEVSALFDSVGVVLTVPEQQLDAVTALSGSGPA
MAV_4652|M.avium_104 AGVTALAKGRFVTAPQLEGVSALFDSVGGVLSVPESQMDAVTALSGSGPA
MAB_4005c|M.abscessus_ATCC_199 AGVSALAKGRFANDEQLSAVAELLESVGTVITVAESQMDTVTAISGSGPA
.**:.** ***.. ::. * ::::** *::*.* *:*:***:******
Mflv_0075|M.gilvum_PYR-GCK YFFLMVEALVDAGVDAGLPRAVATDLAVQTMAGSAAMLLERLDQNAA-G-
Mvan_0837|M.vanbaalenii_PYR-1 YFFLMVEALVDAAVDAGLARSVATDLVVQTMAGSAAMLLERLDESAP-SN
MSMEG_0943|M.smegmatis_MC2_155 YFFLMAEAMVDAGVAVGLSRAVATDLVAQTMAGSAAMLLDRLDQVN----
TH_0955|M.thermoresistible__bu YFFLMAEAMVDAGVAVGLSRAVATGLVAQTMAGSAAMLLDRLDDAAK---
Rv0500|M.tuberculosis_H37Rv YFFLLVEALVDAGVGVGLSRQVATDLAAQTMAGSAAMLLERMEQDQGGAN
Mb0511|M.bovis_AF2122/97 YFFLLVEALVDAGVGVGLSRQVATDLAAQTMAGSAAMLLERMDQDQGGAN
MLBr_02430|M.leprae_Br4923 YFFLLVEALVDAGVAVGLTRQVATELAAQTMAGSAAMLLERMDQDRHSAE
MMAR_0826|M.marinum_M YFLLMVEALVDAGVAAGLSRQVATDLTAQTMAGSAAMLLERMDQDRR-GA
MUL_4570|M.ulcerans_Agy99 YFLLMVEALVDAGVAAGLSRQVATDLTAQTMAGSAAMLLERMDQDRR-GA
MAV_4652|M.avium_104 YFFLLVEALVDAGVAAGLSREVAADLTAQTMAGSAAMLLERMDADRRLGE
MAB_4005c|M.abscessus_ATCC_199 YFFLLVEALVDAGVASGLTRPVATDLVIQTMAGSAAMLLERAESDENRAP
**:*:.**:***.* **.* **: *. ***********:* :
Mflv_0075|M.gilvum_PYR-GCK SASAGPSMDTSAAQLRATVTSPGGTTAAGLRELERGGLRAAVANAVEAAK
Mvan_0837|M.vanbaalenii_PYR-1 DSAAGAFMDTSAAQLRATVTSPGGTTAAGLRELERGGLRAAVANAVEAAK
MSMEG_0943|M.smegmatis_MC2_155 -AAGGAALDTTPAQLRATVTSPAGTTAAGLRELERGGLRAAVANAVEAAK
TH_0955|M.thermoresistible__bu -SAAGAAMDTSPAELRAMVTSPGGTTAAGLRELERGGLRAAVANAVEAAK
Rv0500|M.tuberculosis_H37Rv GELMGLRVDLTASRLRAAVTSPGGTTAAALRELERGGFRMAVDAAVQAAK
Mb0511|M.bovis_AF2122/97 GELMGLRVDLTASRLRAAVTSPGGTTAAALRELERGGFRMAVDAAVQAAK
MLBr_02430|M.leprae_Br4923 VAPLGAQVDVPAAQLRATITSPGGTTAAALRELERGGLRMVVDAAVQAAK
MMAR_0826|M.marinum_M AEPLGLRVDTSPAQLRASVTSPGGTTAAALRELEREGFRGAVDAAVQAAK
MUL_4570|M.ulcerans_Agy99 AEPLGLRLDTSPAQLRASVTSPGGTTAAALRELEREGFRGAVDAAVQAAK
MAV_4652|M.avium_104 AETPGMRVDATATQLRATVTSPGGTTAAGLRELERGGLRAAVDAAVQAAK
MAB_4005c|M.abscessus_ATCC_199 GLQTRTEVDTTAAELRATITSPGGTTAAALRELERGGLRASVYAAVDAAK
:* ..:.*** :***.*****.****** *:* * **:***
Mflv_0075|M.gilvum_PYR-GCK KRSEQLGITTE
Mvan_0837|M.vanbaalenii_PYR-1 TRSEQLGITPE
MSMEG_0943|M.smegmatis_MC2_155 TRSEQLGITSE
TH_0955|M.thermoresistible__bu MRSEQLGITSE
Rv0500|M.tuberculosis_H37Rv SRSEQLRITPE
Mb0511|M.bovis_AF2122/97 SRSEQLRITPE
MLBr_02430|M.leprae_Br4923 IRSEQLRITSE
MMAR_0826|M.marinum_M SRSEQLRITSE
MUL_4570|M.ulcerans_Agy99 SRSEQLRITSE
MAV_4652|M.avium_104 IRSEQLRITSE
MAB_4005c|M.abscessus_ATCC_199 TRSEQLGITSE
***** **.*