For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GWFSAPEYWLGRLALERGTAIIYLIAFVAAAQQFRPLIGEHGMLPVPRYLAGQSFWRTPSIFHFRYSDRV FAGVCWLGAVLSAAVVAGAASFVPLWATMLIWLTLWVLYLSIVNVGQAWYSFGWESLLLETGFLMIFLGN ERTAPPILTLLLARWLLFRVEFGAGLIKMRGDSCWRSLTCLYYHHETQPMPGPLSWFFHHLPKPLHRIEV AGNHFAQLVVPFGLFTPQPAASIAAAIIVVTQLWLVASGNFSWLNWLTILLACSAIDTSSAAALLPMPAQ PALSAPPQWFAGLVVVFTAAVLLLSYWPARNLLSSHQRMNMSFNPFHLVNTYGAFGSICRTRREVVIEGT DESPITEQTVWKAYEFKGKPGDPRRLPRQWAPYHLRLDWLMWFAAISPGYALPWMTPFLNRLLRNDPATL KLLRHNPFPQSPPRYVRAQLYQYRFTTVAELRRDRAWWHRTLIGRYVPPMSLRKVASPPAD*
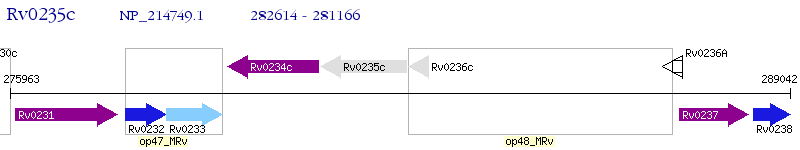
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0235c | - | - | 100% (482) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0240c | - | 0.0 | 100.00% (482) | transmembrane protein |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0493 | - | 0.0 | 81.18% (473) | hypothetical protein MMAR_0493 |
| M. avium 104 | MAV_4924 | - | 0.0 | 79.75% (479) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_1761 | - | 0.0 | 70.00% (430) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1156 | - | 0.0 | 79.37% (475) | hypothetical protein MUL_1156 |
| M. vanbaalenii PYR-1 | Mvan_4797 | - | 0.0 | 71.40% (479) | hypothetical protein Mvan_4797 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0235c|M.tuberculosis_H37Rv --MGWFSAPEYWLGRLALERGTAIIYLIAFVAAAQQFRPLIGEHGMLPVP
Mb0240c|M.bovis_AF2122/97 --MGWFSAPEYWLGRLALERGTAIIYLIAFVAAAQQFRPLIGEHGMLPVP
MMAR_0493|M.marinum_M --MEWFSASEYWLGRLVLERGVAAVYLTAFVASALQFRALIGEHGMLPVP
MUL_1156|M.ulcerans_Agy99 --MEWFSASEYWLGRLVLERGVAAVYLTAFVASALQFRALIGEHGMLPVP
MAV_4924|M.avium_104 --MSWFSAPDYWVGRLVLERGVAVIYLLAFVAAAAQFRALIGEHGMLPIP
Mvan_4797|M.vanbaalenii_PYR-1 MYADWFDAPGYWLAREVLQRGVAAIYLIAFVAAARQFRALIGEHGMLPVP
MSMEG_1761|M.smegmatis_MC2_155 ----------------------------------------------MPIP
:*:*
Rv0235c|M.tuberculosis_H37Rv RYLAGQSFWRTPSIFHFRYSDRVFAGVCWLGAVLSAAVVAGAASFVPLWA
Mb0240c|M.bovis_AF2122/97 RYLAGQSFWRTPSIFHFRYSDRVFAGVCWLGAVLSAAVVAGAASFVPLWA
MMAR_0493|M.marinum_M RYLAHRSFWRTPSVFHFRYSDRLFAGVCWLGAALSAAVVGGTADLVPLWG
MUL_1156|M.ulcerans_Agy99 RYLAHRSFWRTPSVFHFRYSDRLCAGVCWLDAALSAAVVGGTADLVPLWG
MAV_4924|M.avium_104 RFLAGQSWRRTPSIFHLHYSDRLFAALSWFGAALSAAIVAGAADAVPLWA
Mvan_4797|M.vanbaalenii_PYR-1 RFVAQVPFRVAPSVFHLHYSDRFFSAVCWSGAVLATAVAAGGDDLVPLWA
MSMEG_1761|M.smegmatis_MC2_155 RFLARSSFRNSPSIFHLHYSDRFFAAVCWTGAAVAAAMVLGAGDRLPLWA
*::* .: :**:**::****. :.:.* .*.:::*:. * . :***.
Rv0235c|M.tuberculosis_H37Rv TMLIWLTLWVLYLSIVNVGQAWYSFGWESLLLETGFLMIFLGNERTAPPI
Mb0240c|M.bovis_AF2122/97 TMLIWLTLWVLYLSIVNVGQAWYSFGWESLLLETGFLMIFLGNERTAPPI
MMAR_0493|M.marinum_M AMLMWLTLWVLYLSIVNVGQVWYGFGWESLLLETGFLMVFLGNDRVAPPL
MUL_1156|M.ulcerans_Agy99 GMLMWLTLWVLYLSIVNVGQVWYGFGWESLLLETGFLVVFLGNDRVAPPL
MAV_4924|M.avium_104 AMLMWLTLWVLYLSIVNVGQTWYSFGWESLLLETGFLMIFLGNAEVAPPL
Mvan_4797|M.vanbaalenii_PYR-1 ATLAWLALWVLYLSIVNVGQRWYGFGWESLLLEAGFLAMLLGNDDTAPPL
MSMEG_1761|M.smegmatis_MC2_155 AMLVWVLLWVLYLSIVNVGQAWYSFGWESLLLEAGTLAVFLGNDDVAPPL
* *: ************* **.*********:* * ::*** .***:
Rv0235c|M.tuberculosis_H37Rv LTLLLARWLLFRVEFGAGLIKMRGDSCWRSLTCLYYHHETQPMPGPLSWF
Mb0240c|M.bovis_AF2122/97 LTLLLARWLLFRVEFGAGLIKMRGDSCWRSLTCLYYHHETQPMPGPLSWF
MMAR_0493|M.marinum_M VMLWLVRWLLFRVEFGAGLIKMRGDACWRNLTCLYYHHETQPMPGPLSWF
MUL_1156|M.ulcerans_Agy99 VMLWLVRWLLFRVEFGAGLIKMRGDACWRNLTCLYYHHETQPMPGPLSWF
MAV_4924|M.avium_104 LTLWMARLLLFRVEFGAGLIKMRGDPCWRDLTCLYYHHETQPMPGPLSWF
Mvan_4797|M.vanbaalenii_PYR-1 LALWLARWLLFRVEFGAGLIKMRGDPCWRDLSCLDYHHETQPMPGPLSWF
MSMEG_1761|M.smegmatis_MC2_155 LVLWLVRWLLFRVEFGAGMIKMRGDRCWRDLTCLYYHHETQPMPGPLSWF
: * :.* **********:****** ***.*:** ***************
Rv0235c|M.tuberculosis_H37Rv FHHLPKPLHRIEVAGNHFAQLVVPFGLFTPQPAASIAAAIIVVTQLWLVA
Mb0240c|M.bovis_AF2122/97 FHHLPKPLHRIEVAGNHFAQLVVPFGLFTPQPAASIAAAIIVVTQLWLVA
MMAR_0493|M.marinum_M FHRMPRPLHRIEVAGNHFAQLVVPFGLFAPQPIASIAAVIIIVTQLWLVM
MUL_1156|M.ulcerans_Agy99 VHRMPRPLHRIEVAGNHFAQLVVPFGLFAPQPIASIAAVIIIVTQLWLVM
MAV_4924|M.avium_104 FHHLPKPLHRIEVAGNHFAQLIVPFGLFAPQPVAGVAAAIIVVTQLWLVA
Mvan_4797|M.vanbaalenii_PYR-1 FHHLPKPLHRVEVAGNHVAQLVVPFALFAPQPVASVAAGIVIVTQLWLVL
MSMEG_1761|M.smegmatis_MC2_155 FHHLPKPLHRVEVAGNHVAQLVIPFLLFAPQPVASTAAAVVIVTQLWLVA
.*::*:****:******.***::** **:*** *. ** :::*******
Rv0235c|M.tuberculosis_H37Rv SGNFSWLNWLTILLACSAIDTSSAAALLP---MPAQPALSAPPQWFAGLV
Mb0240c|M.bovis_AF2122/97 SGNFSWLNWLTILLACSAIDTSSAAALLP---MPAQPALSAPPQWFAGLV
MMAR_0493|M.marinum_M SGNFSWLNWLTILLACSAIEQSAARKVLP---IPVRAALPPPPTWFAALV
MUL_1156|M.ulcerans_Agy99 SGNFSWLNWLTILLACSAIEQSAARKVLP---IPVRAALPPPPTWFAALV
MAV_4924|M.avium_104 SGNFAWLNWVTILLACSAIDDSSVALLLPKSVLPGQGEPSASPLWFVVVV
Mvan_4797|M.vanbaalenii_PYR-1 SGNFAWLNWLTILLAFSAVDQSVFAVVLP---MSAPPARAGPPPWFAALV
MSMEG_1761|M.smegmatis_MC2_155 SGNFAWLNWITIVLACGVVSDTALSAVVG---VSAAPDVGDPPVWYVAAV
****:****:**:** ..:. : :: :. .* *:. *
Rv0235c|M.tuberculosis_H37Rv VVFTAAVLLLSYWPARNLLSS--HQRMNMSFNPFHLVNTYGAFGSICRTR
Mb0240c|M.bovis_AF2122/97 VVFTAAVLLLSYWPARNLLSS--HQRMNMSFNPFHLVNTYGAFGSICRTR
MMAR_0493|M.marinum_M LIFAATVLVLSYWPARNMLSP--RQRMNMSFNPLHLVNTYGAFGSICRTR
MUL_1156|M.ulcerans_Agy99 LIFAATVLVLSYWPARNMLSPPPRQRMNMSFNPVHLVNTYGAFGSICRTR
MAV_4924|M.avium_104 VAVTAAVLVMSYWPIRNMLSS--RQRMNMSFNSFHLVNTYGAFGSIGRVR
Mvan_4797|M.vanbaalenii_PYR-1 IAGTALTVVLSYWPVRNMLGR--RQQMNASFNPFHLVNTYGAFGSIGRVR
MSMEG_1761|M.smegmatis_MC2_155 VAFTAMVVSLSFWPLRNLVTK--RQRMNMSYNRYHLVNSYGAFGVVGRTR
: :* .: :*:** **:: :*:** *:* ****:***** : *.*
Rv0235c|M.tuberculosis_H37Rv REVVIEGTDESPITEQTVWKAYEFKGKPGDPRRLPRQWAPYHLRLDWLMW
Mb0240c|M.bovis_AF2122/97 REVVIEGTDESPITEQTVWKAYEFKGKPGDPRRLPRQWAPYHLRLDWLMW
MMAR_0493|M.marinum_M REVVIEGTSEAELNAQTVWQEYEFKGKPGRVGRLPRQWAPYHLRLDWLMW
MUL_1156|M.ulcerans_Agy99 REVVVEGTSEAELNAQTVWQEYEFKGKPGRVGRLPRQWAPYHLRLDWLMW
MAV_4924|M.avium_104 REVVIEGTADDDITDQTVWWEYEFKGKPGGVRRLPRQWAPYHLRLDWLMW
Mvan_4797|M.vanbaalenii_PYR-1 YEVVIEGTAEPEPTAQTVWKEYGFKGKPGDLRRMPRQWAPYHLRLDWMMW
MSMEG_1761|M.smegmatis_MC2_155 DEVVIEGTDEPTINDETTWREYEFKGKPGDVRRLPRQFAPYHLRLDWLMW
***:*** : . :*.* * ****** *:***:*********:**
Rv0235c|M.tuberculosis_H37Rv FAAISPGYALPWMTPFLNRLLRNDPATLKLLRHNPFPQSPPRYVRAQLYQ
Mb0240c|M.bovis_AF2122/97 FAAISPGYALPWMTPFLNRLLRNDPATLKLLRHNPFPQSPPRYVRAQLYQ
MMAR_0493|M.marinum_M FAAISPGYALPWMTPLLTRLLRNDRATLRLLRRNPFPDTPPRYVRAQLYQ
MUL_1156|M.ulcerans_Agy99 FAAISPGYALPWMTPLLTRLLRNDRATLRLLRRNPFPDTPPRYVRAQLYQ
MAV_4924|M.avium_104 FAAISPGYAQPWLSPFLQRLLENDRPTLRLLRHNPFPDAPPRYVRARLYE
Mvan_4797|M.vanbaalenii_PYR-1 FAALSPAYARDWFRPLVVRLLRNDAATLRLLGHNPFPDEPPRCIRARLYE
MSMEG_1761|M.smegmatis_MC2_155 FAAISPTYAHPWLRTFLERLLEGDRATLKLLRTNPFPGEPPRYVRARLYR
***:** ** *: .:: ***..* .**:** **** *** :**:**.
Rv0235c|M.tuberculosis_H37Rv YRFTTVAELRRDRAWWHRTLIGRYVPPMSLRKVASPPAD
Mb0240c|M.bovis_AF2122/97 YRFTTVAELRRDRAWWHRTLIGRYVPPMSLRKVASPPAD
MMAR_0493|M.marinum_M YRFTTLQELRRDHAWWHRTLIGKYAAPMSLD--------
MUL_1156|M.ulcerans_Agy99 YPFTTLQELRRHHAWWHRTLIGKYAAPMSLD--------
MAV_4924|M.avium_104 YRFTTPAELRRDRDWWHRTLIGGYVPPLTLSKLTPQDHG
Mvan_4797|M.vanbaalenii_PYR-1 YRFTTWRELRDERRWWHRTLVGEYLPAVALGAGDSPAAR
MSMEG_1761|M.smegmatis_MC2_155 YRYSTWDELRRDHVWWQRTLLGDYVPPVTLRAGHGSAAQ
* ::* *** .: **:***:* * ..::*