For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVVVEKTPTTLPQATPNGAAPWHVRAGAFAIDVLPGLAVAATMALTALTVPPGSAWRWLCACLLGLTIL LLAVNRLLLPTITGWSLGRALTGIRVVRRDGSAIGPWRLLVRDLAHLVDTLSLFVGWLWPLWDSRRRTFA DLLLRTEVRRVEPVQRPAVIRRLTAAVALAAAGACASATAVGAAVVYVNEWQTDHTRAQLATRGPKLVVD VLSYDPETVQRDFERARSLATDRYRPQLSIQQDSVRESGPVRNQYWVTDSAVLSATPAQATMLLFMQGER GTPPNQRYIQSTVRAIFQKSRGQWRLDDLAVVMKPRQPTGEK
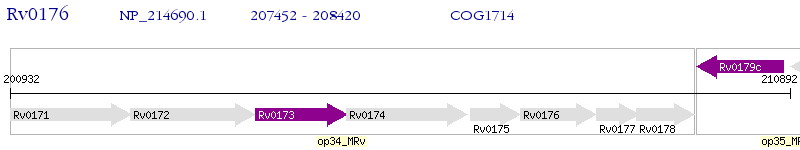
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0176 | - | - | 100% (322) | PROBABLE CONSERVED MCE ASSOCIATED TRANSMEMBRANE PROTEIN |
| M. tuberculosis H37Rv | Rv1078 | pra | 5e-09 | 42.19% (64) | proline-rich antigen |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0182 | - | 0.0 | 99.38% (322) | mce associated transmembrane protein |
| M. gilvum PYR-GCK | Mflv_0704 | - | 2e-75 | 47.09% (327) | RDD domain-containing protein |
| M. leprae Br4923 | MLBr_02596 | - | 2e-98 | 55.17% (319) | hypothetical protein MLBr_02596 |
| M. abscessus ATCC 19977 | MAB_1196 | - | 5e-12 | 31.82% (176) | proline-rich antigen |
| M. marinum M | MMAR_0419 | - | 1e-110 | 59.34% (332) | Mce associated transmembrane protein |
| M. avium 104 | MAV_5008 | - | 1e-103 | 58.33% (324) | RDD family protein |
| M. smegmatis MC2 155 | MSMEG_0141 | - | 2e-77 | 48.40% (312) | mce associated transmembrane protein |
| M. thermoresistible (build 8) | TH_2868 | - | 2e-33 | 45.86% (157) | PUTATIVE RDD domain containing protein |
| M. ulcerans Agy99 | MUL_1069 | - | 1e-107 | 58.43% (332) | Mce associated transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_0141 | - | 4e-83 | 51.08% (323) | RDD domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0704|M.gilvum_PYR-GCK MTVVLDVTETISEPEPTAEVRPASWGARAGALALDILPALGVIATCVVLA
Mvan_0141|M.vanbaalenii_PYR-1 MTAVFDTTPTASADDPTTPMPLASWPARAGAITVDILPGLGVIVTTALLA
TH_2868|M.thermoresistible__bu --------------------------------------------------
MSMEG_0141|M.smegmatis_MC2_155 MSSVLDT-PAAAEDAVIDQAPLASWQARAGAFAIDMLPGAGIIVTLALLA
Rv0176|M.tuberculosis_H37Rv MTVVVEKTPTTL--PQATPNGAAPWHVRAGAFAIDVLPGLAVAATMALTA
Mb0182|M.bovis_AF2122/97 MTVVVEKTPTTL--PQATPNGAAPWHVRAGAFAIDVLPGLAVAATMALTA
MLBr_02596|M.leprae_Br4923 MTVVVAKSQTAAVIPEPLSNRLAPWHLRLVALAVDVLPGLVAVSTMTLVV
MAV_5008|M.avium_104 MTVLVEQTATTG-TPEPVREALAPWHSRVGAFAIDVAPGAAVIATMALVS
MMAR_0419|M.marinum_M MTVVVDKIETTDVVAELPQKVLAPWHLRLGAFAVDVLSGAAVVATLGLVA
MUL_1069|M.ulcerans_Agy99 MTVVVDKIETTDVVAELPQKVLAPWHLRLGAFAVDVLSGAAVVATLGLVA
MAB_1196|M.abscessus_ATCC_1997 ----------MTEVPPPPPQGPGGYPPPPPAAGAPALP------------
Mflv_0704|M.gilvum_PYR-GCK LTAPFDGWVRWLFTVLAAVVFVLTVVNRLVLPAVKGWTLGRALFGIAVRR
Mvan_0141|M.vanbaalenii_PYR-1 LTAPADGWVRWMFIVALAVTFFLMAANRLVLPVATGWTLGRALFGIAVRR
TH_2868|M.thermoresistible__bu --------------------------------------------------
MSMEG_0141|M.smegmatis_MC2_155 WTAPLLGWVWWVYTVAAVLVVGAVLANRVLLPTTRGWTLGRAVVGIRVVK
Rv0176|M.tuberculosis_H37Rv LTVPPGSAWRWLCACLLGLTILLLAVNRLLLPTITGWSLGRALTGIRVVR
Mb0182|M.bovis_AF2122/97 LTVPPGSAWRWLCACLLGLTILLLAVNRLLLPTITGWSLGRALTGIRVVR
MLBr_02596|M.leprae_Br4923 FTVPLRSAWWWLCMAVGGIVILSMLVNRLLLPTIIGWSLGRALCGISVIM
MAV_5008|M.avium_104 FTVPARGVWWWVGVCVLGVVTLLVLVNRLLLPVITGWSLGRALAGIAVTP
MMAR_0419|M.marinum_M ETVPPDSVWWWVCVSLLALVVLVMAANRLLLPPILGWSLGRALCGITVVR
MUL_1069|M.ulcerans_Agy99 ETVPPDSVWWWVCVSLLALVVLVMAANRLLLPPILGWSLGRALCGITVVR
MAB_1196|M.abscessus_ATCC_1997 ------GGNFEIWIRRVGAYIIDVIPVALLSGIGGGIAGGTATTADCVGD
Mflv_0704|M.gilvum_PYR-GCK DDG----GTVGIGRLTVREVAHLLDTLSLFVGWLWPLWDSRRRTFADMVA
Mvan_0141|M.vanbaalenii_PYR-1 ADG----RPVGVLRVTGRELAHLLDTLAVFVGWLWPLWDRRRRTFADLLA
TH_2868|M.thermoresistible__bu --------------------VHQLE---------------RPR-------
MSMEG_0141|M.smegmatis_MC2_155 ADG----GDAAMVRLLLRDLAHVLDTLAVFVGWLWPLWDARNRTFADLLT
Rv0176|M.tuberculosis_H37Rv RDG----SAIGPWRLLVRDLAHLVDTLSLFVGWLWPLWDSRRRTFADLLL
Mb0182|M.bovis_AF2122/97 RDG----SAIGPWRLLVRDLAHLVDTLSLFVGWLWPLWDSRRRTFADLLL
MLBr_02596|M.leprae_Br4923 RDG----VAIGPWRLLLRDLTHLLDTAAVFAGWLWPLWDSRRRTFADILL
MAV_5008|M.avium_104 RTG----AAPGPWRLLLRDLAHLLDTASIVG-WLWPLWDSRRRTFADMLA
MMAR_0419|M.marinum_M RDSHDGDDALGPWLLLVRDLAHLLDTLPLLIGWLWPLWDSRRRTFADLLL
MUL_1069|M.ulcerans_Agy99 RDSHDGDDALGPWLLLVRDLAHLLDTLPLLIGWLWPLWDSRRRTFADLLL
MAB_1196|M.abscessus_ATCC_1997 SFG-----------------------------------------------
Mflv_0704|M.gilvum_PYR-GCK RTRVVRVASPRRD--MRRVVSGVLIATMVAAVAAVGLGYALIFREQRALD
Mvan_0141|M.vanbaalenii_PYR-1 GTEVYRVERPQRD--MRRAVATVLVAAAVASVAAVGLGYALVYRHERAVD
TH_2868|M.thermoresistible__bu -------AAGRRD--PRRWTVVVLVATVLLSVAGAVFGYLTVYRPQQQTD
MSMEG_0141|M.smegmatis_MC2_155 RTEVGRVTPAAGD--VRRLAGKVLLAAVAVCAVAVALGYFAVFRPQEAVH
Rv0176|M.tuberculosis_H37Rv RTEVRRVEPVQRPAVIRRLTAAVALAAAGACASATAVGAAVVYVNEWQTD
Mb0182|M.bovis_AF2122/97 RTEVRRVEPVQRPAVIRRLTAAVALAAAGACASATAVGAAVVYVNEWQTD
MLBr_02596|M.leprae_Br4923 RTEVRCVQAVERQRTIRWWASVALLTAAGVSLGGASVSWAVVYSHDRAID
MAV_5008|M.avium_104 GTEVRRLAPEQVPPHITRWAAAAVLGAAAVCLGGAAMSYGVVYSADRASD
MMAR_0419|M.marinum_M RTEVRYRDPEQRPAHVRQWTVAALLTAAALCVGGSALSYGLVYARDQATD
MUL_1069|M.ulcerans_Agy99 RTEVRYRDPEQRPAHVRQWTVAALLTAAALCVGGSALSYGLVYGRDQATD
MAB_1196|M.abscessus_ATCC_1997 ----------------DPMTGGMSYVSCQPEYTGVGWAAYILFSLAALAF
. : . . ::
Mflv_0704|M.gilvum_PYR-GCK SAREQVAEQGPRIVEQMLSYHVDTLVDDFARAQTLTTDGYRPQLIDQQQA
Mvan_0141|M.vanbaalenii_PYR-1 TAREQVAAQGPRIVEQMLSYGADTIVDDFARAQALTTDGYRPQLISQQQA
TH_2868|M.thermoresistible__bu AARAEIAEQGPRIVEQMLSYQADTIDEDFTRARSLTTERYREQLAAEQQA
MSMEG_0141|M.smegmatis_MC2_155 EAREQIANEGPRIVEQMLSYGVDSFNEDLDKAQTLATDNYRPELVRQQDV
Rv0176|M.tuberculosis_H37Rv HTRAQLATRGPKLVVDVLSYDPETVQRDFERARSLATDRYRPQLSIQQDS
Mb0182|M.bovis_AF2122/97 HTRAQLATRGPKLVVDVLSYDPETVQRDFERARSLATDRYRPQLSIQQDS
MLBr_02596|M.leprae_Br4923 QTRSEIAIQGPKMVAQMLTYNPKSLRDDFTHAQSLASDKYRRQLAAQQDV
MAV_5008|M.avium_104 QTRAQIATQGPKIVAQMLTYDPKSLHDDFARAQSLATDKYRRQLQTQQDV
MMAR_0419|M.marinum_M QTRAEIATQGPKIVAQMLTYDPSTLQEDFARARSLTTDKYREQLAVQQET
MUL_1069|M.ulcerans_Agy99 QTRAEIAAQGPKIVAQMLTYDASTLQEDFARARSLTTDKYREQLAVQQET
MAB_1196|M.abscessus_ATCC_1997 AVWNWGLKQG---------TTGSSIGKGLLGIRVLG--------EATGQP
. .* .:. .: : * :
Mflv_0704|M.gilvum_PYR-GCK VQGAGPVNNEYWAVNSAVLTDPPVTANRVSMLLAMQGQRGTNAEDMKLIT
Mvan_0141|M.vanbaalenii_PYR-1 VQQAGATTNEYWAVNSAVLTDPPVTPDRVSMLLAMQGQRGTDPNDVKFIT
TH_2868|M.thermoresistible__bu VKLTEPTTNDYWAVSSAVLSN---TRDEAAMLLAMQGQRGATPEEQKFIT
MSMEG_0141|M.smegmatis_MC2_155 IRKAGPTTNEYWAVTSAVLSA---TKNDASMLLALQGQRGTNPQDLKFLT
Rv0176|M.tuberculosis_H37Rv VRESGPVRNQYWVTDSAVLSA---TPAQATMLLFMQGERGTPP-NQRYIQ
Mb0182|M.bovis_AF2122/97 VRESGPVRNQYWVTDSAVLSA---TPAQATMLLFMQGERGTPP-NQRYIS
MLBr_02596|M.leprae_Br4923 VKKGHPVINEYWPTAGAIQSA---TRDRATMLLFMQGRRGAAP-GERYIS
MAV_5008|M.avium_104 VQKGNPVINEYWVTNSSIESA---SRDRATMLLFMQGRRGAAP-EVRYIS
MMAR_0419|M.marinum_M VAKGHPVLNEYWVTDSSIQAA---TPDHATMLMFMQGRRGSPP-EIRYIS
MUL_1069|M.ulcerans_Agy99 VAKGHPVLNEYWVTDSSIQAA---TPDHATMLMFMQGRRGSPP-EIRYIS
MAB_1196|M.abscessus_ATCC_1997 IGFGMSVVRQIAHFLDAVICY-------IGFLLPLFTAKRQTIADMLVKT
: .. .: .:: :*: : :
Mflv_0704|M.gilvum_PYR-GCK ATVQVDFEKAADGQWKVANLTVLKRPMMNAGGG
Mvan_0141|M.vanbaalenii_PYR-1 ATVRVDFEKSADGQWRVANLTVLKKPQMNVAGR
TH_2868|M.thermoresistible__bu ATVRVEFKKSPEGQWQVDALTVLKRPYLGEDQP
MSMEG_0141|M.smegmatis_MC2_155 ATVRVDFRKTGD-TWQVDNLTVLKKPNLNGAAQ
Rv0176|M.tuberculosis_H37Rv STVRAIFQKS-RGQWRLDDLAVVMKPRQPTGEK
Mb0182|M.bovis_AF2122/97 ATVRAIFQKS-RGQWRLDDLAVVMKPRQPTGEK
MLBr_02596|M.leprae_Br4923 ATVRVSFAKGEHNHWLVDDLTVLTKPKTTGNGR
MAV_5008|M.avium_104 ATVRVSFAKVGENTWRVDDLTVLTKPKPPGGAK
MMAR_0419|M.marinum_M ATVRVSFAKDGANQWRVDDLTVLTKPKPPGNGK
MUL_1069|M.ulcerans_Agy99 ATVRVSSAKDGANQWRVDDLTVLTKPKPPGNGK
MAB_1196|M.abscessus_ATCC_1997 VVVPK----------------------------
.*