For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTEVPPPPPQGPGGYPPPPPAAGAPALPGGNFEIWIRRVGAYIIDVIPVALLSGIGGGIAGGTATTADCV GDSFGDPMTGGMSYVSCQPEYTGVGWAAYILFSLAALAFAVWNWGLKQGTTGSSIGKGLLGIRVLGEATG QPIGFGMSVVRQIAHFLDAVICYIGFLLPLFTAKRQTIADMLVKTVVVPK
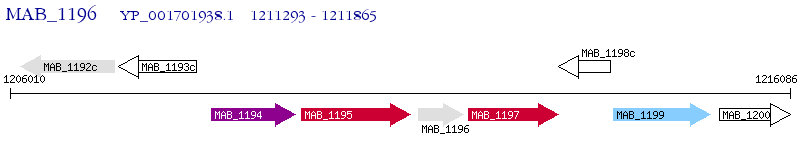
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1196 | - | - | 100% (190) | proline-rich antigen |
| M. abscessus ATCC 19977 | MAB_2634 | - | 5e-05 | 37.84% (74) | hypothetical protein MAB_2634 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1107 | pra | 3e-45 | 50.26% (195) | proline-rich antigen |
| M. gilvum PYR-GCK | Mflv_2045 | - | 4e-27 | 41.29% (155) | RDD domain-containing protein |
| M. tuberculosis H37Rv | Rv1078 | pra | 1e-44 | 49.74% (195) | proline-rich antigen |
| M. leprae Br4923 | MLBr_02395 | Ag36 | 7e-46 | 51.28% (195) | proline rich antigenic protein |
| M. marinum M | MMAR_4389 | pra | 8e-46 | 50.27% (185) | proline-rich antigen, Pra |
| M. avium 104 | MAV_1202 | - | 3e-44 | 45.11% (184) | Pra protein |
| M. smegmatis MC2 155 | MSMEG_5266 | - | 5e-37 | 41.67% (216) | RDD family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0202 | pra | 7e-46 | 50.81% (185) | proline-rich antigen Pra-like protein |
| M. vanbaalenii PYR-1 | Mvan_4672 | - | 5e-34 | 43.98% (166) | RDD domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1196|M.abscessus_ATCC_1997 MTEVPP-------PPPQGP-------------------------------
MSMEG_5266|M.smegmatis_MC2_155 MTQLPPQG---PGPAPYGPHPGGH--------------------------
MLBr_02395|M.leprae_Br4923 MTDQPPPSGSNPTPAPPPPGSSGGYEPSFAPSELGSAYPPPTAPPVGGSY
MAV_1202|M.avium_104 MTDQPPPGGAYP-PPPSSPGSPGG---------------QPTPPPGG--Q
Mb1107|M.bovis_AF2122/97 MTEQPPPGGSYP-PPPPPPGPSGGHEP------------PPAAPPGGSGY
Rv1078|M.tuberculosis_H37Rv MTEQPPPGGSYP-PPPPPPGPSGGHEP------------PPAAPPGGSGY
MMAR_4389|M.marinum_M MTDQPPPGGSYP-PPPPPPGSSGGSEPPAG----GYQAPPPPSAPSGGGY
MUL_0202|M.ulcerans_Agy99 MTDQPPPGGSYP-PPPPPPGSSGGSEPPAG----GYQAPPPPSAPSGGGY
Mflv_2045|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4672|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1196|M.abscessus_ATCC_1997 ------------GGYPPP----------------------------PPAA
MSMEG_5266|M.smegmatis_MC2_155 ---PGPQ-----GGYPPPWQP------------------NPYQTPVPGPY
MLBr_02395|M.leprae_Br4923 PPPPPPG-----GSYPPPPPP------------------GGSYPPPPPST
MAV_1202|M.avium_104 QPPPPPG-----GSYPPPPPP------------------GGSYPPPPPPS
Mb1107|M.bovis_AF2122/97 APPPPPS--SGSGYPPPPPPP-----------------GGGAYPPPPPSA
Rv1078|M.tuberculosis_H37Rv APPPPPS--SGSGYPPPPPPP-----------------GGGAYPPPPPSA
MMAR_4389|M.marinum_M PPPPPPPSPSTSGYPPPPPPPSAPGGSAYPPPPPPPPLEGGYPPPPPPSA
MUL_0202|M.ulcerans_Agy99 PPPPPPPPPSTSGYPPPPPPP---------------PLEGGYPPPPPPSA
Mflv_2045|M.gilvum_PYR-GCK ---------------------------------------------MR---
Mvan_4672|M.vanbaalenii_PYR-1 ---------------------------------------------MTGNL
MAB_1196|M.abscessus_ATCC_1997 G--APALPG--------GNFEIWIRRVGAYIIDVIPVALLSGIGGGIAGG
MSMEG_5266|M.smegmatis_MC2_155 G--PPTVPNP-------QAYTPWFDRVIAFVIDQLPIAVVTVIGYLVVFG
MLBr_02395|M.leprae_Br4923 GAYAPPPPGPAIRSLPKEAYTFWVTRVLAYVIDNIPATVLLGIGMLIQTL
MAV_1202|M.avium_104 GGYAPPPPGPAIRTLPTQDYAPWLTRALAFVIDILPYVVVHGIGTAILVA
Mb1107|M.bovis_AF2122/97 GGYAPPPPGPAIRTMPTESYTPWITRVLAAFIDWAPYVVLVGIGWVIMLV
Rv1078|M.tuberculosis_H37Rv GGYAPPPPGPAIRTMPTESYTPWITRVLAAFIDWAPYVVLVGIGWVIMLV
MMAR_4389|M.marinum_M GGYAPPPPGPAIRALPAESYTPWITRVLAALIDFAPYVVVTGIGWLIMMV
MUL_0202|M.ulcerans_Agy99 GGYAPPPPGPAIRALPAESYTPWITRVLAALIDFAPYVVVTGIGWLIMMV
Mflv_2045|M.gilvum_PYR-GCK -----TAARN--------PHTSWTRRVAAGILDAVPVMLGWGIWESVAIG
Mvan_4672|M.vanbaalenii_PYR-1 SGQPPAMPRR--------VYTSWSARVAAFFVDMVPLGLAWGIWEVVALR
. . . * *. * .:* * : * :
MAB_1196|M.abscessus_ATCC_1997 TATTADCVGDSFGDPMTGGMSYVSCQPEYTGVGWAAYILFSLAALAFAVW
MSMEG_5266|M.smegmatis_MC2_155 ILAGAS-QGSSDGE-ITTGVGIVAAVLIFV---------LSLAPIAYGVW
MLBr_02395|M.leprae_Br4923 TKQEACVTDITQYNVNQYCATQPTGIGMLA------FWFAWLMATAYLVW
MAV_1202|M.avium_104 TQQTACITDVTQYAVNQYCATQNSTLGLVA------QWLASIVGLFYLIW
Mb1107|M.bovis_AF2122/97 TQTSSCVTSISEYDVGQFCVSQPSMIGQLV------QWLLSVGGLAYLVW
Rv1078|M.tuberculosis_H37Rv TQTSSCVTSISEYDVGQFCVSQPSMIGQLV------QWLLSVGGLAYLVW
MMAR_4389|M.marinum_M TTTSSCVTDISQYDIGQYCVSQPSMIGQLA------QWLLSLVGLAYLVW
MUL_0202|M.ulcerans_Agy99 TTTPSCVTDISQYDIGQYCVSQPSMIGQLA------QWLLSLVGLAYLVW
Mflv_2045|M.gilvum_PYR-GCK AASTECVTYDNGGVACTAIGSPAGDVVGVL---------MVFLSVAYLCW
Mvan_4672|M.vanbaalenii_PYR-1 SATLDCVTFDNGGVSCSSGISSVGYLAFAL---------TVAVSVAYLVW
. : *
MAB_1196|M.abscessus_ATCC_1997 NWGLKQGTTGSSIGKGLLGIRVLGEATGQPIGFGMSVVRQIAHFLDAVIC
MSMEG_5266|M.smegmatis_MC2_155 NMGYRQGTTGQSIGKSVMKFKVVSEQTGQPIGFLMSLVRQVAHIVDGLIC
MLBr_02395|M.leprae_Br4923 NYGYRQGATGSSIGKTVMKFKVISEATGQPIGFGMSVVRQLAHFVDAVIC
MAV_1202|M.avium_104 NYGYRQGTTGSSVGKSVMKFKVVSEVTGQPVGFGMSVVRALAHFVDAIIC
Mb1107|M.bovis_AF2122/97 NYGYRQGTTGSSIGKSVLKFKVVSETTGQPIGFGMSVVRQLAHFIDAIIC
Rv1078|M.tuberculosis_H37Rv NYGYRQGTIGSSIGKSVLKFKVVSETTGQPIGFGMSVVRQLAHFIDAIIC
MMAR_4389|M.marinum_M DYGYRQGTTGSSIGKSIMKFKVVSETTGEPLGFGMSVVRQLAHIVDAIIC
MUL_0202|M.ulcerans_Agy99 DYGYRQGTTGSSIGKSIMKFKVVSETTGEPLGFGMSVVRQLAHIVDAVIC
Mflv_2045|M.gilvum_PYR-GCK NFGLRQGRRGASIGKSVMGFRMVDEKTWQAVGFGRSMLRLLVHVVDAVPL
Mvan_4672|M.vanbaalenii_PYR-1 NYGHRQGVSGSSVGKSVLRFQVLDEKTWRPVGFGASVLRQAVHLLDAAVC
: * :** * *:** :: ::::.* * ..:** *::* .*.:*.
MAB_1196|M.abscessus_ATCC_1997 YIGFLLPLFTAKRQTIADMLVKTVVVPK----------------------
MSMEG_5266|M.smegmatis_MC2_155 YVGYLFPLWDAKRQTLADKIMTTVCVPVDRQPPQLSYPPQQQFQQAYAPQ
MLBr_02395|M.leprae_Br4923 CIGFLFPLWDSKRQTLADKIMTTVCLPI----------------------
MAV_1202|M.avium_104 FIGFLFPLWDSKRQTLADKIMTTVCLPLDGSESPPS--------------
Mb1107|M.bovis_AF2122/97 FVGFLFPLWDAKRQTLADKIMTTVCVPI----------------------
Rv1078|M.tuberculosis_H37Rv FVGFLFPLWDAKRQTLADKIMTTVCVPI----------------------
MMAR_4389|M.marinum_M YIGFLFPLWDAKRQTLADKIMTTVCLPL----------------------
MUL_0202|M.ulcerans_Agy99 YIGFLFPLWDAKRQTLADKIMTTVCLPL----------------------
Mflv_2045|M.gilvum_PYR-GCK GIGFLFPLWDRRRQTLADKLMGTVCVPVQGTVLGTRR-------------
Mvan_4672|M.vanbaalenii_PYR-1 FIGFLCPLWDRRRQTLADKLTGTVCVPKRY--------------------
:*:* **: :***:** : ** :*
MAB_1196|M.abscessus_ATCC_1997 --------------------
MSMEG_5266|M.smegmatis_MC2_155 YQQPYPPYPPQNPPGFNEMR
MLBr_02395|M.leprae_Br4923 --------------------
MAV_1202|M.avium_104 --------------------
Mb1107|M.bovis_AF2122/97 --------------------
Rv1078|M.tuberculosis_H37Rv --------------------
MMAR_4389|M.marinum_M --------------------
MUL_0202|M.ulcerans_Agy99 --------------------
Mflv_2045|M.gilvum_PYR-GCK --------------------
Mvan_4672|M.vanbaalenii_PYR-1 --------------------