For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SARCQITGRTVGFGKAVSHSHRRTRRRWPPNIQLKAYYLPSEDRRIKVRVSAQGIKVIDRDGHRGRRRAA RAGSAPAHFARQAGSSLRTAAIL*
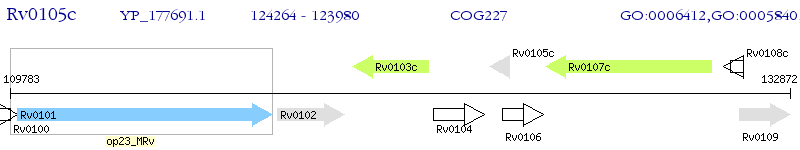
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0105c | rpmB | - | 100% (94) | 50S ribosomal protein L28 |
| M. tuberculosis H37Rv | Rv2058c | rpmB | 5e-24 | 73.02% (63) | 50S ribosomal protein L28 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0108c | rpmB | 2e-50 | 100.00% (94) | 50S ribosomal protein L28 |
| M. gilvum PYR-GCK | Mflv_1312 | - | 1e-24 | 76.19% (63) | ribosomal protein L28 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0334c | rpmB.2 | 5e-27 | 80.95% (63) | 50S ribosomal protein L28-2 |
| M. marinum M | MMAR_0292 | rpmB2 | 1e-27 | 76.00% (75) | 50S ribosomal protein L28 RpmB2 |
| M. avium 104 | MAV_4875 | rpmB | 2e-24 | 77.78% (63) | ribosomal protein L28 |
| M. smegmatis MC2 155 | MSMEG_6068 | rpmB | 2e-24 | 77.78% (63) | ribosomal protein L28 |
| M. thermoresistible (build 8) | TH_2053 | rpmB | 3e-09 | 48.28% (58) | ribosomal protein L28 |
| M. ulcerans Agy99 | MUL_4831 | rpmB2 | 7e-28 | 76.00% (75) | 50S ribosomal protein L28 RpmB2 |
| M. vanbaalenii PYR-1 | Mvan_5489 | rpmB | 8e-26 | 79.37% (63) | 50S ribosomal protein L28 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0105c|M.tuberculosis_H37Rv ----MSARCQITGRTVGFGKAVSHSHRRTRRRWPPNIQLKAYYLPSEDRR
Mb0108c|M.bovis_AF2122/97 ----MSARCQITGRTVGFGKAVSHSHRRTRRRWPPNIQLKAYYLPSEDRR
Mflv_1312|M.gilvum_PYR-GCK ----MSAHCQVTGRKPGFGNAVSHSHRRTRRRWDPNIQRKTYYLPSEGRR
Mvan_5489|M.vanbaalenii_PYR-1 ----MSAHCQVTGRAPGFGNAVSHSHRRTRRRWNPNIQTKTYYLPSEGRR
MAB_0334c|M.abscessus_ATCC_199 MELPLSAHCEVTGRSPGFGNAVSHSHRRTRRRWSPNIQLKTYYLPSEDRR
MSMEG_6068|M.smegmatis_MC2_155 ----MSAHCQVTGRGPGFGNSVSHSHRRTPRRWDPNIQNKTYYLPSEGRR
MMAR_0292|M.marinum_M ----MSARCQVTGRTVSFGNTVSHSHRRSRRRWSPNIQLKTYYLPSENRR
MUL_4831|M.ulcerans_Agy99 ----MSARCQVTGRTVSFGNTVSHSHRRSRRRWSPNIQLKTYYLPSENRR
MAV_4875|M.avium_104 ----MSADCQVTGRRPSFGNAVSHSHRRTPRRWSPNIQLKTYYLPSEGRR
TH_2053|M.thermoresistible__bu ----MAAVCDICGKAPGFGKSVSHSHRRHSRRWNPNIQTVRAVARPGGNK
::* *:: *: .**::******* *** **** . ..:
Rv0105c|M.tuberculosis_H37Rv IKVRVSAQGIKVIDRDGHRGRRRAARAGSAPAHFARQAGSSLRTAAIL
Mb0108c|M.bovis_AF2122/97 IKVRVSAQGIKVIDRDGHRGRRRAARAGSAPAHFARQAGSSLRTAAIL
Mflv_1312|M.gilvum_PYR-GCK ITLRVSTKGIKIIDRDGIES--VVARL--------RREEHKL------
Mvan_5489|M.vanbaalenii_PYR-1 VKLRVSAKGIKVIDRDGIES--VVARL--------RREGQKI------
MAB_0334c|M.abscessus_ATCC_199 IKLRVSTKGIKVIDRDGIEA--VVARL--------RAQGRKF------
MSMEG_6068|M.smegmatis_MC2_155 IRLRVSAKGIKVIDRDGIEA--VVARM--------RARGEKV------
MMAR_0292|M.marinum_M IKLRVSAKGIKVIDRDGIEA--TVARL--------RRAGEQI------
MUL_4831|M.ulcerans_Agy99 IKLRVSAKGIKVIDRDGIEA--TVARL--------RRAGEQI------
MAV_4875|M.avium_104 IRLRVSAKGIKVIDRDGIEA--VVARL--------RRHGQQI------
TH_2053|M.thermoresistible__bu QRMNVCASCLKAG---------KVLRG---------------------
:.*.:. :* . *