For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VESEPLYKLKAEFFKTLAHPARIRILELLVERDRSVGELLSSDVGLESSNLSQQLGVLRRAGVVAARRDG NAMIYSIAAPDIAELLAVARKVLARVLSDRVAVLEDLRAGGSAT
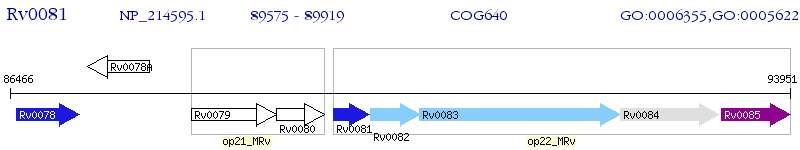
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0081 | - | - | 100% (114) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. tuberculosis H37Rv | Rv1674c | - | 1e-10 | 39.29% (84) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv0324 | - | 5e-10 | 34.41% (93) | ArsR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0084 | - | 6e-57 | 100.00% (114) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_1120 | - | 4e-21 | 47.27% (110) | regulatory protein, ArsR |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2562c | - | 3e-29 | 55.88% (102) | transcriptional regulatory protein ArsR |
| M. marinum M | MMAR_1653 | - | 3e-39 | 75.93% (108) | transcriptional regulatory protein |
| M. avium 104 | MAV_5108 | - | 3e-28 | 54.81% (104) | transcriptional regulator, ArsR family protein |
| M. smegmatis MC2 155 | MSMEG_6451 | - | 2e-18 | 44.44% (108) | transcriptional regulator, ArsR family protein |
| M. thermoresistible (build 8) | TH_2105 | - | 3e-18 | 46.23% (106) | transcriptional regulator, ArsR family protein |
| M. ulcerans Agy99 | MUL_2937 | - | 2e-07 | 40.28% (72) | ArsR-type repressor |
| M. vanbaalenii PYR-1 | Mvan_5692 | - | 4e-21 | 49.09% (110) | regulatory protein, ArsR |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0081|M.tuberculosis_H37Rv -------MESEPLYKLKAEFFKTLAHPARIRILELLVERDR--SVGELLS
Mb0084|M.bovis_AF2122/97 -------MESEPLYKLKAEFFKTLAHPARIRILELLVERDR--SVGELLS
MMAR_1653|M.marinum_M --------MTEPLYKLKAEFFKTLGHPARVRILELLSERDH--TVAELLP
MAB_2562c|M.abscessus_ATCC_199 ------MDARQPLYRMKADFFKTLGHPVRIRVLELLSERDQ--AVSEMLP
MAV_5108|M.avium_104 -----MADSRQPLYRMKAEFFKTLGHPARIRVLELLSSREY--AVSEMLP
Mflv_1120|M.gilvum_PYR-GCK MPHRSPAEQQRPLYEIKANLFKALAHPARIRVLEILATADDPVSVSDMLA
Mvan_5692|M.vanbaalenii_PYR-1 MSDRFPADQERPLYEIKANLFKALAHPARIRVLEILATNRQPTSVSDILA
MSMEG_6451|M.smegmatis_MC2_155 MPHNFLGSPEQPLYEIKANLFKALAHPARIRILEILSASSEPTPVSAILA
TH_2105|M.thermoresistible__bu ----VSDGPERPLYEIKANLFKALAHPARIRVLEILCASGEPTPVSDILA
MUL_2937|M.ulcerans_Agy99 ---------MCTPERICDRIFVALASPVRRKLLEILTGRPLS---AGELS
. .: :* :*. *.* ::**:* . *.
Rv0081|M.tuberculosis_H37Rv SDVGLESSNLSQQLGVLRRAGVVAARRDGNAMIYSIAAPDIAEL---LAV
Mb0084|M.bovis_AF2122/97 SDVGLESSNLSQQLGVLRRAGVVAARRDGNAMIYSIAAPDIAEL---LAV
MMAR_1653|M.marinum_M -EVGLESSNLSQQLGVLRRAGVVSATKDGNTVIYSITSPLIAEL---MAV
MAB_2562c|M.abscessus_ATCC_199 -EVGIEAANLSQQLAVLRRAGLVTARREGLSVLYTLTSPQVAQL---LAT
MAV_5108|M.avium_104 -EVGVEPANLSQQLSILRRAGLVTARREGLSVRYALTSPQVAEL---LVV
Mflv_1120|M.gilvum_PYR-GCK -DTGFEATLLSQHLAVLKKHQVVTARRVGNSVFYELAHPLVSEL---LVI
Mvan_5692|M.vanbaalenii_PYR-1 -STGLEATLLSQHLAVLKRHHVVTARRTGNAVLYEPAHPLISEM---LVI
MSMEG_6451|M.smegmatis_MC2_155 -ETEIEPTLLSQHLAVLKRHHVVSGKRTGNAVFYELAHPKISEL---LVI
TH_2105|M.thermoresistible__bu -DTAIEPTLLSQHLAVLKRHHVVRAQRVGNAVFYELTHPKISEL---LVI
MUL_2937|M.ulcerans_Agy99 DRFELSRPAIAEHLKVLREAGLVSDEAQGRRRIYRLTAEPLAELGEWLHP
.. . ::::* :*:. :* * * : :::: :
Rv0081|M.tuberculosis_H37Rv ARKVLARVLSDRVAVLEDLR------AGGSAT--------
Mb0084|M.bovis_AF2122/97 ARKVLARVLSDRVAVLEDLR------AGGSAT--------
MMAR_1653|M.marinum_M ARRVLTGMLSGQVAMLEDLR------AGVPGADN------
MAB_2562c|M.abscessus_ATCC_199 ARTILTGVVADQAEWLDGAGPVPVGPAEATRARR------
MAV_5108|M.avium_104 ARTILTGVVAGQAETLEEIPESLL--AEPTG---------
Mflv_1120|M.gilvum_PYR-GCK ARTFLADTLAGRRDQLRSIK------ALPPLPGRA-----
Mvan_5692|M.vanbaalenii_PYR-1 ARTFLADTLAGQRDQLKSLE------ALPPLEKRQ-----
MSMEG_6451|M.smegmatis_MC2_155 ARTFLADTLGAQRDQLAAMR------SLPPIGKKPAGKKK
TH_2105|M.thermoresistible__bu ARTFLADTLGAQRDQLETFK------RLPPLGRSQ-----
MUL_2937|M.ulcerans_Agy99 FEKLWREKLSALAEAAEEQQ--------------------
. . :.