For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSVHVAPVTSSTIVVAVDVGKTSAVLSVTDATRHRLLAPSEFAMTGSGTATAAARAMAAAPAAVQVKVGV EAAGHYHRPVLDYRWPEGWEVLELNPAQVAEQRRVQGRRRIKTDVIDLEAITELVLAGYGRPVTYRDVVI GEISAWAVHRSRRVATRSATKNQLLGQLDRAFPGLTLALPDVLATKIGRLIAAEFADPARLAGLGVNRLI RFAATRDLQLRRPVAERLVAAARDALPTRDAVISRRILAADLVLLADLDDQIQSAETALALLLPRSPYAT LTSVPGWAVVRASNYAAALGDPKRWPGPRQIYRASGLSPMQYESAHKRRDGGISREGSVALRRALIDLGI GLWLNEPAAKNYAHALKDRGKPGGIVACALAHRANRIAHALVRDHATYEPARWV
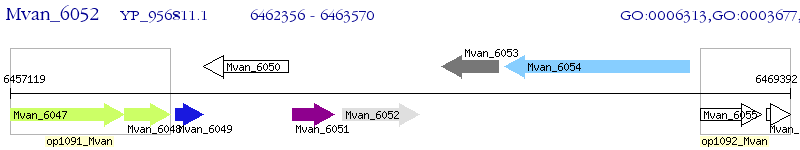
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_6052 | - | - | 100% (404) | transposase IS116/IS110/IS902 family protein |
| M. vanbaalenii PYR-1 | Mvan_4581 | - | 0.0 | 81.68% (404) | transposase IS116/IS110/IS902 family protein |
| M. vanbaalenii PYR-1 | Mvan_0416 | - | 1e-16 | 27.03% (296) | transposase IS116/IS110/IS902 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1870 | - | 0.0 | 88.34% (403) | transposase IS116/IS110/IS902 family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3782 | - | 3e-08 | 24.37% (279) | transposase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4550 | - | 2e-16 | 27.61% (297) | transposase |
| M. thermoresistible (build 8) | TH_3453 | - | 9e-13 | 23.97% (413) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_6052|M.vanbaalenii_PYR-1 ----------------------MSVHVAPVTSSTIVVAVDVGKTSAVLSV
Mflv_1870|M.gilvum_PYR-GCK ----------------------MSVHGAPVTSSTIVVAVDVGKTSAVFSV
MMAR_3782|M.marinum_M -----------------------------MPT--VWAGVDTGKRTHHCVV
MSMEG_4550|M.smegmatis_MC2_155 -----------------------------MAAGQVWAGVDVGKEHHWVCV
TH_3453|M.thermoresistible__bu MEEGVVSKGSGLSRGDKRRNAKLAELRRLVPADHVIVGIDLADKKQAVVV
:.: : ..:* .. *
Mvan_6052|M.vanbaalenii_PYR-1 TDATRHRLLAPSEFAMTGSG-TATAAARAMAAAPAAVQVKVGVEAAGHYH
Mflv_1870|M.gilvum_PYR-GCK TDAARHRLLTPSEFAMNRSG-LIAATTRVMAAVPASGQVKVAVEAAGHYH
MMAR_3782|M.marinum_M LDRNGGVLLS--RPVDNDET-ALLELIEAVVGVAAGREVCWATDLNAGGA
MSMEG_4550|M.smegmatis_MC2_155 VDDSGKVVLS--RRLVNDEQ-PIRELVAEIDELAER--VLWAVDLTTVYA
TH_3453|M.thermoresistible__bu CDHDSQVLARKTVKARAWELGPVLEWAQRVAHKHGFCGVTVGCEPTGHRW
* : . : * . :
Mvan_6052|M.vanbaalenii_PYR-1 RPVLDYRWPEGWEVLELNPAQVAEQRRVQGRRRIKTDVIDLEAITELVLA
Mflv_1870|M.gilvum_PYR-GCK RPVLDHRWPEGWEVVELNPAHVAEQRRVQGRRRIKTDVIDLEAITELVLS
MMAR_3782|M.marinum_M AMLITLLAAHDQQLLYIPGRIVHHAAATYRG-DGKTDAKNARIIADQARM
MSMEG_4550|M.smegmatis_MC2_155 ALLLTVLADAGKTVRYLAGRAVWQASATYRGGEAKTDAKDARVIADQSRM
TH_3453|M.thermoresistible__bu MVLNQLATQRDMTLMCVNPMLVGRAREAEDYTRDKSDDKDAMLIARLVAQ
: . : : * . . *:* : *:
Mvan_6052|M.vanbaalenii_PYR-1 G--YGRPVTYRDVVIGEISAWAVHRSRRVATRSATKNQLLGQLDRAFPGL
Mflv_1870|M.gilvum_PYR-GCK G--HGCSVVDRDVVIGELSAWAAHRSRRVATRSATKNQLLGQLDRAFPGL
MMAR_3782|M.marinum_M R-TDLQPVRRADPIATDLRLLTSRRTDVVCDRVRAINRLRQTLLEYFPPL
MSMEG_4550|M.smegmatis_MC2_155 RGQDLPVLHPNDDLISELRMLTGHRADLVADRTRTINRLRQQLVAVCPAL
TH_3453|M.thermoresistible__bu L--HCYRPERAEETWARLRQLGARRERLVTEATACVQQLRDLLECVWPGV
: : :* * ::* * * :
Mvan_6052|M.vanbaalenii_PYR-1 TLALPDVLATKIGRLIAAEFAD-----PARLAGLGVNRLIRFAATR----
Mflv_1870|M.gilvum_PYR-GCK TLALPDVLATRIGRLVAAEFSD-----PARLAGLGVSRLIRFAATR----
MMAR_3782|M.marinum_M ERAF-DYSKSKAALTLLSRYQT-----PDSLRRIGPTRLATWLKAR----
MSMEG_4550|M.smegmatis_MC2_155 ERAA-QLSQDRGWVVLLARYQR-----PKAIRQSGVSRLTKVLTDA----
TH_3453|M.thermoresistible__bu LEAAADPFGSSNWCAAVAVVLDRCDGRPERLKGRGLARFEAAVVRELGRW
* : : * : * *:
Mvan_6052|M.vanbaalenii_PYR-1 -DLQLRRPVAERLVAAARDALPTR-DAVISRRILAADLVLLADLDDQIQS
Mflv_1870|M.gilvum_PYR-GCK -DLQLRRPVAERLVAAARDALPTR-DAMIARQILAADMNLLADLDDQIQS
MMAR_3782|M.marinum_M -GCRNSATVAQTAVQAAQTQHTTLPTQTVGAQLVPRLAAEITTIDAELAH
MSMEG_4550|M.smegmatis_MC2_155 -GVRNAASIAGAAVAAVKTQTVRLPGEEVAAGLVADLAREVIALDDRIKT
TH_3453|M.thermoresistible__bu GGQRRRRAIIEAVFVALVDPAGVMVQRRGVLERARWVLSDWRAASIRLAE
. : .: . * . .:
Mvan_6052|M.vanbaalenii_PYR-1 AETALALLLPRSP-YATLTSVPGWAVVRASNYAAALG-DPKRWPGPRQIY
Mflv_1870|M.gilvum_PYR-GCK AEAALGELLPRSP-YATLTSVPGWGVVRVSNYAAALG-DPKRWPGPRQIY
MMAR_3782|M.marinum_M VDNEITARFADHDSAEILTSMPGFGPVLAATFLAQIGGNLDGFDTVDRLA
MSMEG_4550|M.smegmatis_MC2_155 TDADIEGRFRRHPLAEVITSMPGMGFRLGAELLAAVG-DPALIGSADQLA
TH_3453|M.thermoresistible__bu VEARMVEVLDELELTELVTSIPGVSAVGAAAMLAETG-DPTRFDSPRALV
.: : : :**:** . : * * : :
Mvan_6052|M.vanbaalenii_PYR-1 RASGLSPMQYESAHKRRDGGISREGSVALRRALIDLGIGLWLNEPAAKN-
Mflv_1870|M.gilvum_PYR-GCK RASGLSPMQYESAHKRRDGGISREGSVALRRALIDLGMGLWLNEPTAKR-
MMAR_3782|M.marinum_M CVAGLAPVPRDSGRISGNLHRPRRFNRRLLRPCYLAALSSLKNSPASRT-
MSMEG_4550|M.smegmatis_MC2_155 AWAGLAPVSRDSRKRTGRLHTPKRYSRRLRRVMYMSALTAIRCDPDSKA-
TH_3453|M.thermoresistible__bu KHAGLCPRDNASGTFNGRARISRRGRPRLRLAAWRATWGALQHNPVMLAR
:**.* * .:. * .*
Mvan_6052|M.vanbaalenii_PYR-1 --YAHALKDRGKPGGIVACALAHRANRIAHALVRDHATYEPARWV-----
Mflv_1870|M.gilvum_PYR-GCK --YAHGLKDRGKPGGVVACALAHRANRIAHALVRDHATYEPTRWA-----
MMAR_3782|M.marinum_M --FYDRKRAEGKSHKQALIALARRRINVIWAMLRDHTHYHEPDLVNTPLA
MSMEG_4550|M.smegmatis_MC2_155 --YYQRKRDEGKRPIPATICLARRRTNVLYALIRDNRTWQPESPPNIAAA
TH_3453|M.thermoresistible__bu YRYLTTRDDNKLTDGQARAAIAAALLRWLHVVVTQRVRWDGAIAAAQVMP
: . . .:* . .:: :. :.
Mvan_6052|M.vanbaalenii_PYR-1 ---
Mflv_1870|M.gilvum_PYR-GCK ---
MMAR_3782|M.marinum_M A--
MSMEG_4550|M.smegmatis_MC2_155 ---
TH_3453|M.thermoresistible__bu VAA