For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVHGAPVTSSTIVVAVDVGKTSAVFSVTDAARHRLLTPSEFAMNRSGLIAATTRVMAAVPASGQVKVAVE AAGHYHRPVLDHRWPEGWEVVELNPAHVAEQRRVQGRRRIKTDVIDLEAITELVLSGHGCSVVDRDVVIG ELSAWAAHRSRRVATRSATKNQLLGQLDRAFPGLTLALPDVLATRIGRLVAAEFSDPARLAGLGVSRLIR FAATRDLQLRRPVAERLVAAARDALPTRDAMIARQILAADMNLLADLDDQIQSAEAALGELLPRSPYATL TSVPGWGVVRVSNYAAALGDPKRWPGPRQIYRASGLSPMQYESAHKRRDGGISREGSVALRRALIDLGMG LWLNEPTAKRYAHGLKDRGKPGGVVACALAHRANRIAHALVRDHATYEPTRWA*
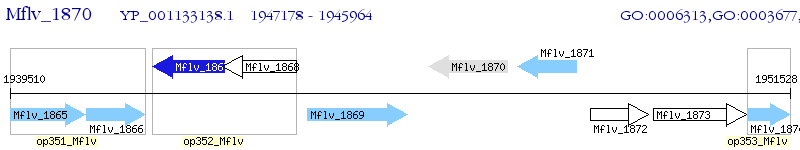
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1870 | - | - | 100% (404) | transposase IS116/IS110/IS902 family protein |
| M. gilvum PYR-GCK | Mflv_4199 | - | 0.0 | 87.53% (361) | transposase IS116/IS110/IS902 family protein |
| M. gilvum PYR-GCK | Mflv_4200 | - | 2e-18 | 27.80% (295) | transposase IS116/IS110/IS902 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3782 | - | 8e-08 | 23.72% (312) | transposase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4550 | - | 4e-18 | 27.91% (301) | transposase |
| M. thermoresistible (build 8) | TH_3017 | - | 4e-14 | 26.47% (272) | PUTATIVE transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_6052 | - | 0.0 | 88.34% (403) | transposase IS116/IS110/IS902 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3782|M.marinum_M ---------MPTVWAGVDTGKRTHHCVVLDRNGGVLLS--RPVDNDETAL
TH_3017|M.thermoresistible__bu --------------------------VVIDAQGKRLLS--QRVANDEATL
MSMEG_4550|M.smegmatis_MC2_155 -------MAAGQVWAGVDVGKEHHWVCVVDDSGKVVLS--RRLVNDEQPI
Mflv_1870|M.gilvum_PYR-GCK MSVHGAPVTSSTIVVAVDVGKTSAVFSVTDAARHRLLTPSEFAMNRSGLI
Mvan_6052|M.vanbaalenii_PYR-1 MSVHVAPVTSSTIVVAVDVGKTSAVLSVTDATRHRLLAPSEFAMTGSGTA
* * :*: . . .
MMAR_3782|M.marinum_M LELIEAVVGVAAGREVCWATDLNAGGAAMLITLLAAHDQQLLYIPGRIVH
TH_3017|M.thermoresistible__bu LKLISTVATMADGGEVTWAIDLNAGGAALLITLLIAADQRLLYIPGRTVH
MSMEG_4550|M.smegmatis_MC2_155 RELVAEIDELAER--VLWAVDLTTVYAALLLTVLADAGKTVRYLAGRAVW
Mflv_1870|M.gilvum_PYR-GCK AATTRVMAAVPASGQVKVAVEAAGHYHRPVLDHRWPEGWEVVELNPAHVA
Mvan_6052|M.vanbaalenii_PYR-1 TAAARAMAAAPAAVQVKVGVEAAGHYHRPVLDYRWPEGWEVLELNPAQVA
: . * . : :: . : : *
MMAR_3782|M.marinum_M HAAATYRG-DGKTDAKNARIIADQARMR-TDLQPVRRADPIATDLRLLTS
TH_3017|M.thermoresistible__bu HASGSYRG-EGKTDAKDAAVIADQARMR-RDLQPLRPGDDITVELRILTS
MSMEG_4550|M.smegmatis_MC2_155 QASATYRGGEAKTDAKDARVIADQSRMRGQDLPVLHPNDDLISELRMLTG
Mflv_1870|M.gilvum_PYR-GCK EQRRVQGRRRIKTDVIDLEAITELVLSG--HGCSVVDRDVVIGELSAWAA
Mvan_6052|M.vanbaalenii_PYR-1 EQRRVQGRRRIKTDVIDLEAITELVLAG--YGRPVTYRDVVIGEISAWAV
. ***. : *:: : * : :: :
MMAR_3782|M.marinum_M RRTDVVCDRVRAINRLRQTLLEYFPPLERAF-DYSKSKAALTLLSRYQTP
TH_3017|M.thermoresistible__bu RRTDLVADRTRTINRLRAQLLEYFPALERAF-DYSTSKAALLLLTGYQTP
MSMEG_4550|M.smegmatis_MC2_155 HRADLVADRTRTINRLRQQLVAVCPALERAA-QLSQDRGWVVLLARYQRP
Mflv_1870|M.gilvum_PYR-GCK HRSRRVATRSATKNQLLGQLDRAFPGLTLALPDVLATRIGRLVAAEFSDP
Mvan_6052|M.vanbaalenii_PYR-1 HRSRRVATRSATKNQLLGQLDRAFPGLTLALPDVLATKIGRLIAAEFADP
:*: *. * : *:* * * * * : : : : : *
MMAR_3782|M.marinum_M DSLRRIGPTRLATWLKARGCRNSATVAQTAVQAAQTQHTTLPTQTVGAQL
TH_3017|M.thermoresistible__bu DGLRRAGAARLAVWLGKRKARNADAVAAKALQAAHAQHTVIPGQQLAAAM
MSMEG_4550|M.smegmatis_MC2_155 KAIRQSGVSRLTKVLTDAGVRNAASIAGAAVAAVKTQTVRLPGEEVAAGL
Mflv_1870|M.gilvum_PYR-GCK ARLAGLGVSRLIRFAATRDLQLRRPVAERLVAAARDALPTR-DAMIARQI
Mvan_6052|M.vanbaalenii_PYR-1 ARLAGLGVNRLIRFAATRDLQLRRPVAERLVAAARDALPTR-DAVISRRI
: * ** : .:* : *.: :. :
MMAR_3782|M.marinum_M VPRLAAEITTIDAELAHVDNEITARFADHDSAEILTSMPGFGPVLAATFL
TH_3017|M.thermoresistible__bu VARLAKEVMALDTEIGDTDAMIEERFHRHRHAEIIVSMSGFGVTLGAEFL
MSMEG_4550|M.smegmatis_MC2_155 VADLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGMGFRLGAELL
Mflv_1870|M.gilvum_PYR-GCK LAADMNLLADLDDQIQSAEAALGELLPRSPYATLTS-VPGWGVVRVSNYA
Mvan_6052|M.vanbaalenii_PYR-1 LAADLVLLADLDDQIQSAETALALLLPRSPYATLTS-VPGWAVVRASNYA
:. : :* .: .: : : * : :.* . :
MMAR_3782|M.marinum_M AQIGGNLDGFDTVDRLACVAGLAPVPRDSGRISGNLHRPRRFNRRLLRPC
TH_3017|M.thermoresistible__bu AATGGDMSAFDSVDRLAGVAGLAPVPRDSGRISGNLQRPRRYNRRLLRVC
MSMEG_4550|M.smegmatis_MC2_155 AAVG-DPALIGSADQLAAWAGLAPVSRDSRKRTGRLHTPKRYSRRLRRVM
Mflv_1870|M.gilvum_PYR-GCK AALG-DPKRWPGPRQIYRASGLSPMQYESAHKRRDGGISREGSVALRRAL
Mvan_6052|M.vanbaalenii_PYR-1 AALG-DPKRWPGPRQIYRASGLSPMQYESAHKRRDGGISREGSVALRRAL
* * : :: :**:*: :* : .:. . * *
MMAR_3782|M.marinum_M YLAALSSLKNSPASRTFYDRKRAEGKSHKQALIALARRRINVIWAMLRDH
TH_3017|M.thermoresistible__bu YLSALSSIRSDPASRTYYNRKRAEGKRHSQAVLALARRRLNVLWAMLRDH
MSMEG_4550|M.smegmatis_MC2_155 YMSALTAIRCDPDSKAYYQRKRDEGKRPIPATICLARRRTNVLYALIRDN
Mflv_1870|M.gilvum_PYR-GCK IDLGMGLWLNEPTAKRYAHGLKDRGKPGGVVACALAHRANRIAHALVRDH
Mvan_6052|M.vanbaalenii_PYR-1 IDLGIGLWLNEPAAKNYAHALKDRGKPGGIVACALAHRANRIAHALVRDH
.: .* :: : . : .** . .**:* .: *::**:
MMAR_3782|M.marinum_M THYHEPDLVNTPLAA
TH_3017|M.thermoresistible__bu TTYQPTTPTAAAAA-
MSMEG_4550|M.smegmatis_MC2_155 RTWQPESPPNIAAA-
Mflv_1870|M.gilvum_PYR-GCK ATYEPTRWA------
Mvan_6052|M.vanbaalenii_PYR-1 ATYEPARWV------
:.