For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRKQAHTAKAAEAVAGTPWSITDMVLDIIDRELAVPAPERGAALIAVRDSRLIVDVVSDPRPGESVSYWH SDQLRRRLSDYLTDNPTRRYVGTVHSHPGGYAEPSGPDHQAFANMLATNRAIRDAIFPIVVQAPRDGLGS VLRLGDKHLADLPHGTFAGYSAHPTDAGLVVRPAPIHVVPAGAHAAVVADALTRHLGQPVTVRWGEPLTV SGTAWLTAQFMIDTRTIAGVALGPSYPMTPPMVWAAESASPVFSSWPISATGDHLAGAVSAVLQHQTPPV AAVRAGIRERLSVHLPNQIDAHVLLIGAGSVGSNAAEMLIRSGVKRLTVVDFDTVEPANLSRTVYGSGDL GQLKTAALADRLTAIAADVEITQLTCPLQELTGEVLDTVDLAFLASDDMAGEGWLNHELYVRAIPSVSVK VFAGAEGAELAYVLPARNTACLRCMMGAVGSGDRGDADYGTGRINGSPALGPDIAAAAARGVKVALALTQ TDGPLADWLDELVSRRLTYFLSSNVAGWTYTEFARAGSLPFDGLWLTAPGRPECEICGMERVSDTQPAHV AAFTTSPPQGIGDVDTDDHRHGEDN*
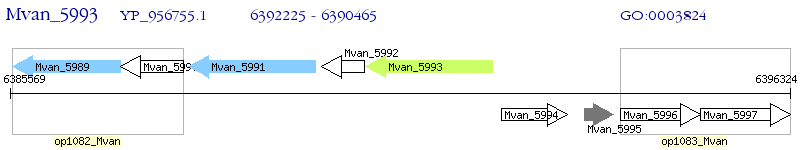
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5993 | - | - | 100% (586) | UBA/THIF-type NAD/FAD binding protein |
| M. vanbaalenii PYR-1 | Mvan_1797 | - | 1e-06 | 34.41% (93) | molybdopterin biosynthesis-like protein MoeZ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3143 | moeB2 | 4e-08 | 30.00% (150) | molybdenum cofactor biosynthesis protein MoeB |
| M. gilvum PYR-GCK | Mflv_4670 | - | 2e-06 | 28.33% (120) | molybdopterin biosynthesis-like protein MoeZ |
| M. tuberculosis H37Rv | Rv3116 | moeB2 | 4e-08 | 30.00% (150) | molybdenum cofactor biosynthesis protein MoeB |
| M. leprae Br4923 | MLBr_00817 | moeZ | 2e-07 | 36.11% (72) | molybdopterin biosynthesis-like protein MoeZ |
| M. abscessus ATCC 19977 | MAB_3525c | - | 8e-07 | 36.99% (73) | molybdenum cofactor biosynthesis protein MoeB1 |
| M. marinum M | MMAR_1352 | moeB1 | 1e-07 | 36.56% (93) | molybdenum cofactor biosynthesis protein MoeB1 |
| M. avium 104 | MAV_4153 | - | 1e-07 | 38.36% (73) | molybdopterin biosynthesis-like protein MoeZ |
| M. smegmatis MC2 155 | MSMEG_1937 | - | 2e-06 | 35.48% (93) | molybdopterin biosynthesis-like protein MoeZ |
| M. thermoresistible (build 8) | TH_0450 | moeZ | 2e-08 | 41.10% (73) | PROBABLE MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN MOEB1 |
| M. ulcerans Agy99 | MUL_2527 | moeB1 | 6e-08 | 36.56% (93) | molybdopterin biosynthesis-like protein MoeZ |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1352|M.marinum_M ---MSTTALPPLVAPAATLSREEVARYSRHLIIPDLGVDGQKR------L
MUL_2527|M.ulcerans_Agy99 ---MSTTALPPLVAPAATLSREEVARYSRHLIIPDLGVDGQKR------L
MLBr_00817|M.leprae_Br4923 -MSTSSTSLPPLVEPAGQLSRDEVIRYSRHLIIPDIGVDGQMR------L
Mflv_4670|M.gilvum_PYR-GCK ----MSTPLPPLVEPAAELTKEEVARYSRHLIIPDLGLDGQKR------L
MSMEG_1937|M.smegmatis_MC2_155 MSANLDVSLPPLVEPADELTREEVARYSRHLIIPDLGLDGQKR------L
MAB_3525c|M.abscessus_ATCC_199 -----MPTLPPLVRPAAELTREEVARYSRHLIIPDLGVDGQKR------L
TH_0450|M.thermoresistible__bu ----VSSPLPPLVEPAAELTREEVARYSRHLIIPDVGVEGQKR------L
MAV_4153|M.avium_104 ----MSTPLPPLVAPADQLTADEMARYSRQLIIPGLGVDGQKR------L
Mb3143|M.bovis_AF2122/97 ----MTEALIPAPS-QISLTRDEVRRYSRHLIIPDIGVNGQQR------L
Rv3116|M.tuberculosis_H37Rv ----MTEALIPAPS-QISLTRDEVRRYSRHLIIPDIGVNGQQR------L
Mvan_5993|M.vanbaalenii_PYR-1 --MMRKQAHTAKAAEAVAGTPWSITDMVLDIIDRELAVPAPERGAALIAV
. . : .: .:* :.: . * :
MMAR_1352|M.marinum_M KNARVLVIGAGG--------------LGSPTLLYLAAAG----VGTIGIV
MUL_2527|M.ulcerans_Agy99 KNARVLVIGAGG--------------LGSPTLLYLAAAG----VGTIGIV
MLBr_00817|M.leprae_Br4923 KSARVLVIGAGG--------------LGAPVLLYLAAAG----VGTIGII
Mflv_4670|M.gilvum_PYR-GCK KNARVLVIGAGG--------------LGSPTLLYLAAAG----VGTIGIV
MSMEG_1937|M.smegmatis_MC2_155 KNAKVLVIGAGG--------------LGSPTLLYLAAAG----VGTIGIV
MAB_3525c|M.abscessus_ATCC_199 KNAKVLVIGAGG--------------LGSPTLLYLAAAG----VGTIGIV
TH_0450|M.thermoresistible__bu KNARVLVIGAGG--------------LGSPALLYLAAAG----VGTLGVI
MAV_4153|M.avium_104 KNARVLVIGAGG--------------LGAPTLLYLAAAG----VGTIGIV
Mb3143|M.bovis_AF2122/97 KDARVLCIGAGG--------------LGSPALLYLAAAG----VGTIGII
Rv3116|M.tuberculosis_H37Rv KDARVLCIGAGG--------------LGSPALLYLAAAG----VGTIGII
Mvan_5993|M.vanbaalenii_PYR-1 RDSRLIVDVVSDPRPGESVSYWHSDQLRRRLSDYLTDNPTRRYVGTVHSH
:.:::: ... * **: ***:
MMAR_1352|M.marinum_M DFDVVDESNLQRQIIHGVADVGRSK--------AQSARDSIVAINPLIEV
MUL_2527|M.ulcerans_Agy99 DFDVVDESNLQRQIIHGVADVGRSK--------AQSARDSIVAINPLIEV
MLBr_00817|M.leprae_Br4923 DFDVVDESNLQRQIIHGVADVGRSK--------AQSARDSIVAINPLVQV
Mflv_4670|M.gilvum_PYR-GCK EFDVVDESNLQRQIIHGQSDIGRPK--------AQSAKDSVLEVNPLVTV
MSMEG_1937|M.smegmatis_MC2_155 EFDVVDESNLQRQIIHGQSDIGRSK--------AQSARDSIAEINPLVTV
MAB_3525c|M.abscessus_ATCC_199 EFDVVDESNLQRQIIHGQSDIGRSK--------AQSARDSVLEINPLVEV
TH_0450|M.thermoresistible__bu DFDVVDESNLQRQVIHGQADVGRPK--------AESARDTIRAINPFVDV
MAV_4153|M.avium_104 EFDAVEESNLQRQIIHGVADVGRSK--------AASARDSIAAINPLVDV
Mb3143|M.bovis_AF2122/97 DGDHVDESNLQRQIIHGTSDVGRPK--------VESAAEAVAEINPHVRV
Rv3116|M.tuberculosis_H37Rv DGDHVDESNLQRQIIHGTSDVGRPK--------VESAAEAVAEINPHVRV
Mvan_5993|M.vanbaalenii_PYR-1 PGGYAEPSGPDHQAFANMLATNRAIRDAIFPIVVQAPRDGLGSVLRLGDK
. .: *. ::* : . .*. . :. : : :
MMAR_1352|M.marinum_M RLHEFRLESDNAVDLFGHYDLIIDGTDNFATRYLVNDAAVLAGKP-----
MUL_2527|M.ulcerans_Agy99 RLHEFRLESDNAVDLFGHYDLIIDGTDNFATRYLVNDAAVLAGKP-----
MLBr_00817|M.leprae_Br4923 RLHEFRLESSNVVNLFKQYDLIVDGTDNFATRYLINDAAVLAKKP-----
Mflv_4670|M.gilvum_PYR-GCK NLHEFRLEPDNAVELFEQYDLILDGTDNFATRYLVNDAAVLAGKP-----
MSMEG_1937|M.smegmatis_MC2_155 NLHEFRLEPDNAVDLFGQYDLILDGTDNFATRYLVNDAAVLAGKP-----
MAB_3525c|M.abscessus_ATCC_199 RLHELRLDPENAVELFAQYDLILDGTDNFATRYLVNDAAVLAHKP-----
TH_0450|M.thermoresistible__bu RLHQFRLDRSNAVDLFSEYDLILDGTDNFATRYLVNDAAVLAGKP-----
MAV_4153|M.avium_104 RLHEFRLDASNAVELFGHYDLIVDGTDNFATRYLINDAAVLAGKP-----
Mb3143|M.bovis_AF2122/97 TQYREMLTHDNALEIFGDHDLIVDGTDNFTTRYLINDAAVLAGKP-----
Rv3116|M.tuberculosis_H37Rv TQYREMLTHDNALEIFGDHDLIVDGTDNFTTRYLINDAAVLAGKP-----
Mvan_5993|M.vanbaalenii_PYR-1 HLADLPHGTFAGYSAHPTDAGLVVRPAPIHVVPAGAHAAVVADALTRHLG
. . :: . : . .***:*
MMAR_1352|M.marinum_M ----YVWGSIYRFEG---QVSVFWEDAPGGLGLNYRDLYPEPPPP-----
MUL_2527|M.ulcerans_Agy99 ----YVWGSIYRFEG---QVSVFWEDAPGGLGLNYRDLYPEPPPP-----
MLBr_00817|M.leprae_Br4923 ----YVWGSLYRFEG---QVSVFWEDAPDGLGLNYRDLYLEPPPP-----
Mflv_4670|M.gilvum_PYR-GCK ----YVWGSIYRFEG---QVSIFWEDAPNGLGLNYRDLYPEPPPP-----
MSMEG_1937|M.smegmatis_MC2_155 ----YVWGSIYRFEG---QVSVFWEDAPDGLGLNYRDLYPEPPPP-----
MAB_3525c|M.abscessus_ATCC_199 ----YVWGSIYRFEG---QVSVFWEDAPDGLGLNYRDLYPEPPPP-----
TH_0450|M.thermoresistible__bu ----YVWGSIYRFEG---QVSVFWEDAPNGQGLNYRDLYPEPPPP-----
MAV_4153|M.avium_104 ----YVWGSIYRFEG---QASVCWEDAPDGRGLNYRDLYPEPPPP-----
Mb3143|M.bovis_AF2122/97 ----YVWGSIYRFNG---QTSVFWP----GRGPCYRCLHPAPPPP-----
Rv3116|M.tuberculosis_H37Rv ----YVWGSIYRFNG---QTSVFWP----GRGPCYRCLHPAPPPP-----
Mvan_5993|M.vanbaalenii_PYR-1 QPVTVRWGEPLTVSGTAWLTAQFMIDTRTIAGVALGPSYPMTPPMVWAAE
**. ..* .: * : .**
MMAR_1352|M.marinum_M ------GMVPSCAEGGVLGIICASIASVMGTEAVKLITGIGETLLGRLM-
MUL_2527|M.ulcerans_Agy99 ------GMVPSCAEGGVLGIICASIASVMGTEAVKLITGIGETLLGRLM-
MLBr_00817|M.leprae_Br4923 ------GMVPSCAEGGVLGIVCASIASVMGTEAIKLITGIGAPLLGRLM-
Mflv_4670|M.gilvum_PYR-GCK ------GMVPSCAEGGVLGILCASIASVMGTEAIKLITGIGDPLLGRLM-
MSMEG_1937|M.smegmatis_MC2_155 ------GMVPSCAEGGVLGILCSSIASIMGTEAIKLITGIGEPLLGRLM-
MAB_3525c|M.abscessus_ATCC_199 ------GMVPSCAEGGVLGILCASIASVMGTEAIKLITGIGDSLLGRLM-
TH_0450|M.thermoresistible__bu ------GMVPSCAEGGVLGILPATIGAIQGTEAIKLITGIGETLLGQLL-
MAV_4153|M.avium_104 ------GAVPSCAEGGVLGVVCAAIASVMSTEVIKLITGIGESLLGRLM-
Mb3143|M.bovis_AF2122/97 ------GLVPSCAEGGVLGAICATIASIQVTEVLKLLTGVGTPLVGRLL-
Rv3116|M.tuberculosis_H37Rv ------GLVPSCAEGGVLGAICATIASIQVTEVLKLLTGVGTPLVGRLL-
Mvan_5993|M.vanbaalenii_PYR-1 SASPVFSSWPISATGDHLAGAVSAVLQHQTPPVAAVRAGIRERLSVHLPN
. * .* *. *. ::: . . : :*: * :*
MMAR_1352|M.marinum_M IYDALEMTYRTIRIRKDP------ATPKITALIDYEQFCG----------
MUL_2527|M.ulcerans_Agy99 IYDALEMTYRTIRIRKDP------ATPKITALIDYEQFCG----------
MLBr_00817|M.leprae_Br4923 IYNALEMSYRRIRIHKDP------SRPKITELTDYQQFCG----------
Mflv_4670|M.gilvum_PYR-GCK VYDALDMSYRTIKIRKDP------STPKITELIDYEAFCG----------
MSMEG_1937|M.smegmatis_MC2_155 VYDALDMTYRTIRIRKDP------STPKITELIDYEEFCG----------
MAB_3525c|M.abscessus_ATCC_199 VYDALDMTYRTIKIRKDP------ATPKITELIDYEAFCG----------
TH_0450|M.thermoresistible__bu IYDALEMSYRKIRIRKDP------STPAITELIDYEEFCG----------
MAV_4153|M.avium_104 IYDALEMSYRTIAIRRDPCD---ASRPAITTLVDYEQLCGA---------
Mb3143|M.bovis_AF2122/97 MYEALDATYHQIRIAKNPDCAICGDAPTITELVDDSVSCA----------
Rv3116|M.tuberculosis_H37Rv MYEALDATYHQIRIAKNPDCAICGDAPTITELVDDSVSCA----------
Mvan_5993|M.vanbaalenii_PYR-1 QIDAHVLLIGAGSVGSNAAEMLIRSGVKRLTVVDFDTVEPANLSRTVYGS
:* : :. : * .
MMAR_1352|M.marinum_M ---------VVSDDVARAASDSTITPQELRAMIDSGKKLALIDVREQVEW
MUL_2527|M.ulcerans_Agy99 ---------VVSDDVARAASDSTITPQELRAMINSGKKLALIDVREQVEW
MLBr_00817|M.leprae_Br4923 ---------VVSDDAAQVATGSIVTPRELRELLDSGKKLALIDVREPVEW
Mflv_4670|M.gilvum_PYR-GCK ---------VVSDAAAEAAADSTITPRELRELLDSGKPLALIDVREPVEW
MSMEG_1937|M.smegmatis_MC2_155 ---------VVSDEAAAAAADSTVTPRELKELLDSGKPLALIDVRERVEW
MAB_3525c|M.abscessus_ATCC_199 ---------VISDEASAAVADSTITPRELRELLDTDKKVALIDVREPVEW
TH_0450|M.thermoresistible__bu ---------VVSDAAAEAAADSTVTPRELRELLDAGK-VALVDVREPVEW
MAV_4153|M.avium_104 -------APAASTDAATGGAEAAITPRQLRELLDSGAKLALIDVREPVEF
Mb3143|M.bovis_AF2122/97 -------------STQSVDPELVISCDELRTKQQSDQNFLLVDVREPAEF
Rv3116|M.tuberculosis_H37Rv -------------STQSVDPELVISCDELRTKQQSDQNFLLVDVREPAEF
Mvan_5993|M.vanbaalenii_PYR-1 GDLGQLKTAALADRLTAIAADVEITQLTCPLQELTGEVLDTVDLAFLASD
. :: :. . :*: ..
MMAR_1352|M.marinum_M DIV------------HIDGAQLIPKSLISTGEGLAKLPQDRTTVLYCKTG
MUL_2527|M.ulcerans_Agy99 DIV------------HIDGAQLIPKSLISTGEGLAKLPQDRTTVLYCKTG
MLBr_00817|M.leprae_Br4923 DIV------------HIDGAQLIPRSLINSGEGLAKLPQDRMSVLYCKTG
Mflv_4670|M.gilvum_PYR-GCK DIN------------RIEGAELIPKGGFESGEALSKLDNDRTPVFYCKTG
MSMEG_1937|M.smegmatis_MC2_155 DIN------------HIEGAELIPKPTFESGAALAKLPTDRTPVFYCKTG
MAB_3525c|M.abscessus_ATCC_199 DIV------------HIDGAELVPKSTLESGDGLAKLPQDRQAVLYCKTG
TH_0450|M.thermoresistible__bu EIN------------RIEGAELIPKSEIESGVGLAKLPMDRAPVLYCKTG
MAV_4153|M.avium_104 DIV------------HLDGAQLIPQSSINSGEGLAKLPADRMPVLYCKTG
Mb3143|M.bovis_AF2122/97 DIA------------HIPGSILIPKGEIGSAAGLAQLPLDKEIVLYCKSG
Rv3116|M.tuberculosis_H37Rv DIA------------HIPGSILIPKGEIGSAAGLAQLPLDKEIVLYCKSG
Mvan_5993|M.vanbaalenii_PYR-1 DMAGEGWLNHELYVRAIPSVSVKVFAGAEGAELAYVLPARNTACLRCMMG
:: : . : . * . : * *
MMAR_1352|M.marinum_M ----------------VRSAEALAALKKAGFAD-------AVHLQGGIVA
MUL_2527|M.ulcerans_Agy99 ----------------VRSAEALAALKKAGFAD-------AAHLQGGIVA
MLBr_00817|M.leprae_Br4923 ----------------VRSAETLATVKKAGFSD-------AVHLQGGIVA
Mflv_4670|M.gilvum_PYR-GCK ----------------IRSAEVLAIVKKAGFAD-------AMHVQGGIVA
MSMEG_1937|M.smegmatis_MC2_155 ----------------VRSAEVLAIAKKAGFSD-------AVHLQGGIVA
MAB_3525c|M.abscessus_ATCC_199 ----------------IRSAEALVAVKKAGFSD-------AVHLQGGIAA
TH_0450|M.thermoresistible__bu ----------------VRSAEALAAVKRAGFAD-------AMHLQGGIVA
MAV_4153|M.avium_104 ----------------VRSAQALAVVRQAGFSD-------AVHLQGGIVA
Mb3143|M.bovis_AF2122/97 ----------------IRSAQALTTLKAAGLHN-------VKHLDGGIAE
Rv3116|M.tuberculosis_H37Rv ----------------IRSAQALTTLKAAGLHN-------VKHLDGGIAE
Mvan_5993|M.vanbaalenii_PYR-1 AVGSGDRGDADYGTGRINGSPALGPDIAAAAARGVKVALALTQTDGPLAD
:..: .* *. : :* :.
MMAR_1352|M.marinum_M WANQMQPDMVMY--------------------------------------
MUL_2527|M.ulcerans_Agy99 WANQMQPDMVMY--------------------------------------
MLBr_00817|M.leprae_Br4923 WAKQVQPDMVIY--------------------------------------
Mflv_4670|M.gilvum_PYR-GCK WGKQLEPDMVMY--------------------------------------
MSMEG_1937|M.smegmatis_MC2_155 WAKQLEPDMVMY--------------------------------------
MAB_3525c|M.abscessus_ATCC_199 WAKQVDPDMVMY--------------------------------------
TH_0450|M.thermoresistible__bu WAKQLEPDMVMY--------------------------------------
MAV_4153|M.avium_104 WAQQMQPDMILY--------------------------------------
Mb3143|M.bovis_AF2122/97 WTRTIDSSLLVY--------------------------------------
Rv3116|M.tuberculosis_H37Rv WTRTIDSSLLVY--------------------------------------
Mvan_5993|M.vanbaalenii_PYR-1 WLDELVSRRLTYFLSSNVAGWTYTEFARAGSLPFDGLWLTAPGRPECEIC
* : . : *
MMAR_1352|M.marinum_M --------------------------------------
MUL_2527|M.ulcerans_Agy99 --------------------------------------
MLBr_00817|M.leprae_Br4923 --------------------------------------
Mflv_4670|M.gilvum_PYR-GCK --------------------------------------
MSMEG_1937|M.smegmatis_MC2_155 --------------------------------------
MAB_3525c|M.abscessus_ATCC_199 --------------------------------------
TH_0450|M.thermoresistible__bu --------------------------------------
MAV_4153|M.avium_104 --------------------------------------
Mb3143|M.bovis_AF2122/97 --------------------------------------
Rv3116|M.tuberculosis_H37Rv --------------------------------------
Mvan_5993|M.vanbaalenii_PYR-1 GMERVSDTQPAHVAAFTTSPPQGIGDVDTDDHRHGEDN