For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRQKRTVAVGAAAVAVAAVIASCSNTAGNEASSTASPAPVQTSAATPAASHNQADMMFTRHMIPHHQQAI EMSDMIIAKQGIDPRVVDLAEQIKAAQGPEIAQMQGWLDEWGMPGNMPPGHGDMPGGPMMPGMPGMEGMP GMEGMMSPADMQALQNAQGVEASKLFLSQMIRHHEGAITMAENEIENGQSPDVVELAKTIASSQRTEIDT MNQILKSL
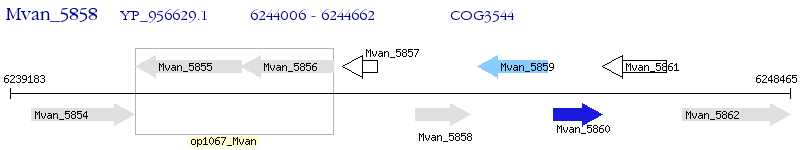
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5858 | - | - | 100% (218) | hypothetical protein Mvan_5858 |
| M. vanbaalenii PYR-1 | Mvan_3676 | - | 1e-74 | 58.78% (245) | hypothetical protein Mvan_3676 |
| M. vanbaalenii PYR-1 | Mvan_4447 | - | 2e-44 | 44.29% (219) | hypothetical protein Mvan_4447 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3889 | - | 1e-62 | 56.44% (225) | hypothetical protein Mflv_3889 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4543c | - | 3e-33 | 38.64% (220) | hypothetical protein MAB_4543c |
| M. marinum M | MMAR_5232 | - | 4e-05 | 37.10% (62) | hypothetical protein MMAR_5232 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5017 | - | 1e-52 | 50.23% (217) | lipoprotein |
| M. thermoresistible (build 8) | TH_3901 | - | 3e-35 | 40.20% (204) | PUTATIVE putative protein of unknown function DUF305 |
| M. ulcerans Agy99 | MUL_4307 | - | 3e-05 | 37.10% (62) | hypothetical protein MUL_4307 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5858|M.vanbaalenii_PYR-1 -MRQKRTVAVGAAAVAVAAVIAS-CSN-----TAGNEASSTASPAPVQT-
Mflv_3889|M.gilvum_PYR-GCK --MQHRGFRFGFAAIAASALVVSACSNGST--DSSSDSATTSAAASATS-
MSMEG_5017|M.smegmatis_MC2_155 ---MKKVLAGGISVFALSFVVAG-CGS-----DTTAEQAQTTSAAAAPS-
MAB_4543c|M.abscessus_ATCC_199 --MRNRTRTAILAGAAAAVLILSACSSDNKGDDAKSEHSGHSGMSGMTTT
TH_3901|M.thermoresistible__bu MTTTTARIVALLAALATAAFLAG-CSS-----TDTGSTPETSTPETGTD-
MMAR_5232|M.marinum_M ----------MQPGGDMSALLAQ---------AQQMQQKLLEAQQQLAN-
MUL_4307|M.ulcerans_Agy99 ----------MQPGGDMSALLAQ---------AQQMQQKLLEAQQQLAN-
. : .: .
Mvan_5858|M.vanbaalenii_PYR-1 ---SAATPAASHNQADMMFTRHMIPHHQQAIEMSDMIIAKQGIDPRVVDL
Mflv_3889|M.gilvum_PYR-GCK ---SVASGATAHNGSDVMFAQGMIPHHQQAIEMSDMLLGKQDIDPEVVSL
MSMEG_5017|M.smegmatis_MC2_155 ---TTQQ-GRPHNDADVMFAQHMIPHHQQAVEMSDIILGKPDIDPRVTDL
MAB_4543c|M.abscessus_ATCC_199 LDTNAAAPVSDHNDADVTFAQQMIMHHQQAVEMSKLTEGR-TANPAVLDL
TH_3901|M.thermoresistible__bu ---TTQGQAAAHNDADIAFVTDMIPHHQQALELSALVPDR-SENTQLIEL
MMAR_5232|M.marinum_M --------SEVHGQAGGGLVQVAVRGSGELVGVT--------IDPKVVDR
MUL_4307|M.ulcerans_Agy99 --------SEVHGQAGGGLVQVAVRGSGELVGVT--------IDPKVVDR
*. :. :. : : : :: :. : .
Mvan_5858|M.vanbaalenii_PYR-1 AEQIKAAQGPEIAQMQGWLDEWGMPGNMPPGHGDMPGGPM----MPGMPG
Mflv_3889|M.gilvum_PYR-GCK AEQIKSAQGPEIEQMQGWLQEWGV-GSTPPGDGTMPGHHMPDHQMPHGGG
MSMEG_5017|M.smegmatis_MC2_155 ADQIQAAQGPEIDQMQSWLKQWGN----PP--------------MGDHEG
MAB_4543c|M.abscessus_ATCC_199 AKNIETAQQPEIDTFTGWLKQWNQP--------------------LMPEG
TH_3901|M.thermoresistible__bu AAGIAAAQDPEIQAMRALLVQWNADDPDGEHGDHAVP--------GMPPG
MMAR_5232|M.marinum_M DD-IETLQDLIVGAMRDASQQVTK------------------------MA
MUL_4307|M.ulcerans_Agy99 DD-IETLQDLIVGAMRDASQQVTK------------------------MA
* : * : : : .
Mvan_5858|M.vanbaalenii_PYR-1 MEGMPGM----EGMMSPADMQALQNAQGVEASKLFLSQMIRHHEGAITMA
Mflv_3889|M.gilvum_PYR-GCK MGPMPGMPAGGHGMMSQADMTALQNAQGPEAGRLFLTQMITHHEGAIMMA
MSMEG_5017|M.smegmatis_MC2_155 MGDHGGM----AGMVSDADMTALRDAQGVDAAKLFLTHMIAHHEGAIDMA
MAB_4543c|M.abscessus_ATCC_199 HDPAGHMP----GMVDAPVLDRMKTLNGEVFDQLWLQSMIAHHQGAIAMS
TH_3901|M.thermoresistible__bu MGPHHDMP----GMVDPATMARLESLRGTEFDRLWLESMIAHHEGAVEMA
MMAR_5232|M.marinum_M QERLGALA----GAMRPPGAPGAPGAPG--------APGMPPRPGAPGMP
MUL_4307|M.ulcerans_Agy99 QERLGALA----GAMRPPGAPGAPGAPG--------APGMPPRPGAPGMP
: * : . * : : ** *.
Mvan_5858|M.vanbaalenii_PYR-1 ENEIENGQSPDVVELAKTIASSQRTEIDTMNQILKSL
Mflv_3889|M.gilvum_PYR-GCK QREIESGQFPAAVDMARSIISSQQSEIDQMRAMLAG-
MSMEG_5017|M.smegmatis_MC2_155 QEEVNEGRFEDAVQMARTIVITQEQEIKDMRQVLDSL
MAB_4543c|M.abscessus_ATCC_199 NAELSGGQYPAAKQLAQQIIDNQQPEIDTMQGLLKG-
TH_3901|M.thermoresistible__bu QTELAEGENVDARTLAQQIIDAQQAEIDQMRQMQEAS
MMAR_5232|M.marinum_M GMPGAPG----------------APGMPGMPGAPGV-
MUL_4307|M.ulcerans_Agy99 GMPGAPG----------------APGMPGMPGAPGV-
* : *