For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQPIPVIALTGYLGAGKTTLLNHVLRSPDARIGVVINDFGELNVDAGLVTGQVDEPASIAGGCICCLPDE GGLDSALARLADPKLRLDAIIVEASGLADPVAVSRIIRFSGVDGVRPGGVVDVLDAATHFDTVDRDATPP ARYGAATLVVVNKLDRIPVRDRADALVRIESRVRELNPHAHVVGATAGRIDPALLYDVAGDAETLGQLTF RELWSDAEAQAHPHVHDHDHSHADSVTVTGEGCVDPAAVIDLLEAPPDGVYRMKGTVAVRYRSSSRCYSV NVVGPSVHIAVAPPRAVANSLVAIGMGLDVEAVRERMRSALTPVQGAASAHGLRRLQRYRKLSI
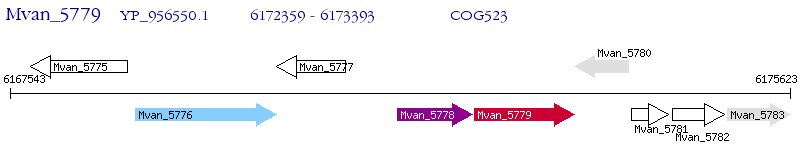
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5779 | - | - | 100% (344) | cobalamin synthesis protein, P47K |
| M. vanbaalenii PYR-1 | Mvan_5321 | - | 5e-23 | 27.73% (357) | cobalamin synthesis protein, P47K |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1052 | - | 5e-76 | 95.83% (144) | cobalamin synthesis protein, P47K |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3425c | - | 5e-51 | 38.27% (324) | putative cobalamin synthesis protein |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1123 | - | 5e-48 | 39.64% (333) | cobalamin synthesis protein |
| M. thermoresistible (build 8) | TH_4457 | - | 1e-131 | 67.94% (340) | PUTATIVE cobalamin synthesis protein, P47K |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5779|M.vanbaalenii_PYR-1 ----------------MQPIPVIALTGYLGAGKTTLLNHVLR-SPDARIG
Mflv_1052|M.gilvum_PYR-GCK MGPDLQIDSGGVASPAVQPIPVIALTGYLGAGKTTLLNHVLR-SPDARIG
TH_4457|M.thermoresistible__bu ----------------VEPIPVIALTGYLGAGKTSLLNHVLR-TPGARIG
MAB_3425c|M.abscessus_ATCC_199 ------------------MIPVTIVSGFLGAGKTTLLNNLLRNTLGVRIG
MSMEG_1123|M.smegmatis_MC2_155 -------------MRTTGSVPVLALAGYLGAGKTTLLNHLLRNSRGVRIG
:** ::*:******:***::** : ..***
Mvan_5779|M.vanbaalenii_PYR-1 VVINDFGELNVDAGLVTGQVDEPASIAGGCICCLPDEGGLDSALARLADP
Mflv_1052|M.gilvum_PYR-GCK VVINDFGELNVDAGLVTGQVDEPASIAGGCICCLPDEGGLDVALERLADP
TH_4457|M.thermoresistible__bu VVINDFGDINVDASMVTGQIDEPASIAGGCICCLPDDGELDVALAKLADP
MAB_3425c|M.abscessus_ATCC_199 VVVNDFGAINIDAMLVAGQVDSTMTMSNGCLCCTADDDELGGMLSALAGR
MSMEG_1123|M.smegmatis_MC2_155 ALVNDFGAVNIDAMLVAGQVDAMAGLTNGCICCAVDADEAGEMLGKLAAV
.::**** :*:** :*:**:* ::.**:** * . . * **
Mvan_5779|M.vanbaalenii_PYR-1 KLRLDAIIVEASGLADPVAVSRIIRFSGVDGVRP--GGVVDVLDAATHFD
Mflv_1052|M.gilvum_PYR-GCK KLRLDAIIVEASGLADPVAVSRIIRFSGVDGIRP--GGVVDVLDAATHFD
TH_4457|M.thermoresistible__bu RLRLDAIIIEASGVAEPASIARMIGFSGVDGIRD--GGVVDVIDAANHFD
MAB_3425c|M.abscessus_ATCC_199 D--IDAIVIEASGLAEPLPLVRMVLAATAEDRRIGYGGLVTVVDTAQYED
MSMEG_1123|M.smegmatis_MC2_155 KPRLDLIVVEASGLAEPAALARTI--AMADDTRYHYAGLVLVVD-----A
:* *::****:*:* .: * : : .:. * .*:* *:*
Mvan_5779|M.vanbaalenii_PYR-1 TVDRDATPPARYGAATLVVVNKLDRIPVRDRADALVRIESRVRELNPHAH
Mflv_1052|M.gilvum_PYR-GCK TIDRDDAPPARYGPP-----------------------------------
TH_4457|M.thermoresistible__bu TVDRGDQPPARYGAATLVVVNKLDQIPEQERPALLRRVEDRVRERNPRAH
MAB_3425c|M.abscessus_ATCC_199 VAVRHPEIAQHLDLADLVVLNKIDLADAGDRTHLR----AVVKERNPRAS
MSMEG_1123|M.smegmatis_MC2_155 VESTRAELGNNVQVADLVVVNKADQVSDAELEAVR----AKITALNDRVP
. . .
Mvan_5779|M.vanbaalenii_PYR-1 VVGATAGRIDPALLYDVAGDAETLGQLTFRELWSDAEAQAHPHVHDHDHS
Mflv_1052|M.gilvum_PYR-GCK ------------------------------PWWSSTSSTGY---------
TH_4457|M.thermoresistible__bu IVGTTGGRIDPALLYDVASAQDETGQLSFRDLFIEPSDHGAHG--DHDHV
MAB_3425c|M.abscessus_ATCC_199 IVETTGADVDPRLLFDKKERPEPVGQLSLEDLIREDHSHEH-----HAHH
MSMEG_1123|M.smegmatis_MC2_155 VLPTEFGRVDPELLVDPPQQVPRVAQLSLDELLWETYDHDH-----HEHA
.
Mvan_5779|M.vanbaalenii_PYR-1 HADSVTVTGEGCVDPAAVIDLLEAPPDGVYRMKGTVAVRYRSSSRCYSVN
Mflv_1052|M.gilvum_PYR-GCK --------------------------------------RPRSVPRRWSAS
TH_4457|M.thermoresistible__bu HADSVTVTSTGTVDPVRLVELLEDPPARVYRLKGTVAVRQRSNPRHYLIN
MAB_3425c|M.abscessus_ATCC_199 HYETVEFATEQPLHPRRLIEFLDSQPAGVYRIKGLVRLGD----RTLAVH
MSMEG_1123|M.smegmatis_MC2_155 TFQSVAFTSDVPLDPRRFMDLLKNRPAGMYRAKGFVDFGDAGEGRKFVLQ
*
Mvan_5779|M.vanbaalenii_PYR-1 VVGPSVHIAVA---PPRAVANSLVAIGMGLDVEAVRERMRSALTPVQGAA
Mflv_1052|M.gilvum_PYR-GCK RAGSAR--------------------------------------------
TH_4457|M.thermoresistible__bu AVGSAVHVTTR---RSRVSENSLVAIGMGLDIDAVRTRLSTALQPFDGPT
MAB_3425c|M.abscessus_ATCC_199 TVGDHISFDKVGRASDFSSGASLVLIGTHLDGDTITERLRTLIDDPPDPD
MSMEG_1123|M.smegmatis_MC2_155 LVGGSLRFERRRWDRGEARRTSLVLIGTDIDEAAALSALQGCAT--AEPA
.*
Mvan_5779|M.vanbaalenii_PYR-1 SAHGLRRLQRYRKLSI
Mflv_1052|M.gilvum_PYR-GCK ----------------
TH_4457|M.thermoresistible__bu SAAALRRLQRYQAGVG
MAB_3425c|M.abscessus_ATCC_199 RERSMLVLGKYLR---
MSMEG_1123|M.smegmatis_MC2_155 DKQAILGVLRYTD---